pyrrolysine
May be freely published and quoted by journals
Name of the committee
The IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN) and Nomenclature Committee of IUBMB (NC-IUBMB) work closely together. In order to acknowledge this, it was agreed that "Joint Committees on Biochemical Nomenclature" should be used as an umbrella term for the two committees.
Enzyme nomenclature
Accepted name
There has been considerable discussion and correspondence on the name by which an enzyme is listed in the EC list. For each enzyme in the EC list, the entry provides two names. The systematic name describes the chemical reaction catalysed, but is often too lengthy for general use. The second name, by which the enzyme is normally described, has been variously called the "Recommended name" or (in the 1992 printed version of the list) "Name" or (in some supplements) "Reaction", or, since 2004, the "Common name". However some of the names are not the ones in most common use. It has now been agreed to use the term 'Accepted name' with the intention to make it easier for editors to enforce its usage.
The "other names" by which enzymes are described in the literature are listed and can be found by the search function on the websites.
Partial EC numbers Often the available experimental evidence about an enzyme is insufficient for a definitive EC number to be assigned. However for purposes such as gene annotation and metabolic pathways an identifier is needed for the enzyme. In many such cases it is possible to decide on the enzyme class and subclass, and in databases such as EMBL Nucleotide Sequence Database, KEGG and UniProt, it has become the practice to use a partial EC number, in which a hyphen indicates the missing subclass or sub-subclass. At the suggestion of Green and Karp, the letter 'n' will be used, such as EC 2.3.4.n, when the enzyme has been characterized, but no EC number has been allocated.
New portal for submission of new enzymes and corrections In addition to the current versions of the Enzyme List already on the web (https://www.qmul.ac.uk/sbcs/iubmb/enzyme), a new MySQL database, ExplorEnz (http://www.enzyme-database.org) has been set up by Keith F. Tipton, Andrew McDonald, and Sinéad Boyce. It is synchronized with the enzyme list on the JCBN website. Since the beginning of 2009, Sinéad Boyce is no longer the contact point for suggestions for new enzymes, and a new procedure has been set up to deal with pending submissions. The ExplorEnz website serves as a portal for proposed new enzymes in the EC list. A task force of enzyme experts has been convened, to undertake the assignment of new EC-numbers.
Other nomenclature recommendations
Use of gene symbols for gene products
Gene symbols are used increasingly in papers to represent the proteins that are the gene products. The committee strongly discourages this practice, which can lead to much confusion. Conventionally, in print the genes are written in italics and the product names in roman characters. However this distinction is lost in most forms of electronic communication that use plain text, such as ASCII. Protein names in general, and enzyme names in particular, provide some indication of what the protein does, this is not the case with gene names. Moreover names using the relative molecular mass on a gel etc. are uninformative as the values tend to vary between species.
Guidelines for naming proteins Ambiguities regarding gene/protein names are a major problem in the literature and it is even worse in the sequence databases which tend to propagate the confusion. A preliminary set of rules, for the construction and usage of names of proteins, has been agreed by the UniProt Consortium, comprising Swiss Institute of Bioinformatics (SIB), Geneva, Switzerland, the European Bioinformatics Institute (EBI), Hinxton, UK, and the Protein Information Resource (PIR), Washington DC, USA. The preliminary document is available at https://www.qmul.ac.uk/sbcs/iubmb/proteinName.html. These recommendations have been endorsed by the Nomenclature Committee.
Lipid classification A classification of lipids has been published by the Lipid MAPS (Metabolites and Pathways Strategy) consortium ( http://www.lipidmaps.org/data/classification/LM_classification_exp.php). This classification has also been adopted by the KEGG database. It divides lipids into eight categories: fatty acyl compounds, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides. Although this is not a recognized IUPAC nomenclature document it is based on, and extends, the published JCBN recommendations.
Metabolic charts Don Nicholson has produced the 22nd edition of his Metabolic Pathways chart, and 35 mini-maps (each in three formats, svg, pdf and gif). There are also 5 Animaps, which are animated web pages, the latest of which illustrates the mechanism of mitochondrial respiratory chain. A new "Inborn Errors of Metabolism" chart has now been produced, which contains links to the most common inherited diseases involving deficiencies of enzymes. These maps and animations are made available to the biochemical community, for example for teaching purposes, and the copyright has been transferred to IUBMB. They have been put on the web at http://www.iubmb-nicholson.org. The large charts and posters have been produced in printed form by Sigma-Aldrich (http://www.sigmaaldrich.com/).
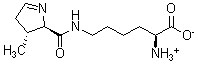
Pyrrolysine Pyrrolysine, found in proteins of certain archaea, is the 22nd naturally occurring amino acid known to be encoded by a specific triplet code (UAG, usually a stop codon) in messenger RNA. In order to representing pyrrolysine in sequence databases, the three-letter code 'Pyl' and the one-letter code 'O' are recommended. The three-letter abbreviation is included in the Enzyme List for EC 6.1.1.25, lysine-tRNA ligase.

pyrrolysine
It is recommended that a comma be used between the symbols for two different substituents on the same functional group, in particular, on the N-terminal amino group of a peptide, e.g., Bn,Me-Trp-Gly-... Similarly, two substituents on a side-chain amino group are indicated within parentheses and are separated by a comma, e.g. ...-Lys(Ac,Me)-Ser-... Identical substituents are indicated by a subscript index, as usual, e.g. Me2-Lys-... However, care should be taken when specifying substituents on the nitrogen atom of a peptide bond within a peptide chain. It is recommended that such a substituent be placed within parentheses on the left-hand side of the amino-acid symbol, e.g. Lys-(Me)Ala-... Note that the position of the hyphen indicates the position of the methyl group, i.e. that it is attached to the amino group of the alanyl residue, whereas Lys(Me)-Ala-... indicates that the methyl group is attached to N6 of the lysyl residue.
Note: Lys-Me-Ala, which is analogous to the form used for substituents on the N-terminal amino group, is not an alternative option because it places the methyl group between the two amino-acid residues.
IUPAC publications of interest to Biochemistry and Molecular Biology Nomenclature of Inorganic Chemistry: IUPAC Recommendations 2005, (Red Book) Royal Society of Chemistry, 2005. Edited by N. G. Connelly and T. Damhus (with R. M. Hartshorn and A. T. Hutton) [ISBN 0-85404-438-8].
Graphical representation of stereochemical configuration - (IUPAC recommendations 2006). Brecher, J. Pure Appl. Chem., 78 (2006) 1897-1970.
Quantities, Units and Symbols in Physical Chemistry, (The Green Book) 3rd edition, RSC Publishing, 2007. Prepared by E.R. Cohen, T. Cvitaš, J.G. Frey, B. Holmström, K. Kuchitsu, R. Marquardt, I. Mills, F. Pavese, M. Quack, J. Stohner, H.L. Strauss, M. Takami and A.J. Thor. [ISBN 9780-85404-433-7]
Graphical representation standards for chemical structure diagrams (IUPAC Recommendations 2008). Brecher, J. Pure Appl. Chem., 80 (2008) 277-410.
References Enzyme Nomenclature 1992, Academic Press, San Diego, California, 1992 prepared by E.C. Webb [ISBN 0-12-227164-5 (hardback), 0-12-227165-3 (paperback)]
Green, M.L. and Karp, P.D. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acid Res. 33 (2005) 4035-4039 [PMID 16034025].
Fahy, E., Subramaniam, S., Brown, H.A., Glass, C.K., Merrill, A.H., Murphy, R.C., Raetz, C.R.H., Russell, D.W., Seyama, Y., Shaw, W., Shimizu, T., Spener, F., van Meer, G., VanNieuwenhze, M.S., White, S.H., Witztum, J.L. and Dennis, E.A. A comprehensive classification system for lipids. J. Lipid Res. 46 (2005) 839-861 [PMID 15722563].
Harvey, D.J. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom. Rev. 18 (1999) 349-450.
Krzycki, J.A. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 8 (2005) 706-12. [PMID 16256420]