Enzyme Nomenclature. Recommendations 1992
Continued from EC 3.4.16 to EC 3.4.20
EC 3.4.21
EC 3.4.21 Serine endopeptidases
Contents
See separate file for EC 3.4.21.51 to EC 3.4.21.100 and EC 3.4.21.101 to EC 3.4.21.122.
EC 3.4.21.1 chymotrypsin
EC 3.4.21.2 chymotrypsin C
EC 3.4.21.3 metridin
EC 3.4.21.4 trypsin
EC 3.4.21.5 thrombin
EC 3.4.21.6 coagulation factor Xa
EC 3.4.21.7 plasmin
EC 3.4.21.8 now covered by EC 3.4.21.34, EC 3.4.21.35
EC 3.4.21.9 enteropeptidase
EC 3.4.21.10 acrosin
EC 3.4.21.11 now covered by EC 3.4.21.36, EC 3.4.21.37
EC 3.4.21.12 α-lytic endopeptidase
EC 3.4.21.13 now EC 3.4.16.1
EC 3.4.21.14 now covered by EC 3.4.21.62 to EC 3.4.21.65, EC 3.4.21.67
EC 3.4.21.15 now EC 3.4.21.63
EC 3.4.21.16 deleted
EC 3.4.21.17 deleted
EC 3.4.21.18 deleted
EC 3.4.21.19 glutamyl endopeptidase
EC 3.4.21.20 cathepsin G
EC 3.4.21.21 coagulation factor VIIa
EC 3.4.21.22 coagulation factor IXa
EC 3.4.21.23 deleted
EC 3.4.21.24 deleted
EC 3.4.21.25 cucumisin
EC 3.4.21.26 prolyl oligopeptidase
EC 3.4.21.27 coagulation factor XIa
EC 3.4.21.28 deleted, included in EC 3.4.21.74
EC 3.4.21.29 deleted, included in EC 3.4.21.74
EC 3.4.21.30 deleted, included in EC 3.4.21.74
EC 3.4.21.31 now covered by EC 3.4.21.68, EC 3.4.21.73
EC 3.4.21.32 brachyurin
EC 3.4.21.33 deleted
EC 3.4.21.34 plasma kallikrein
EC 3.4.21.35 tissue kallikrein
EC 3.4.21.36 pancreatic elastase
EC 3.4.21.37 leukocyte elastase
EC 3.4.21.38 coagulation factor XIIa
EC 3.4.21.39 chymase
EC 3.4.21.40 deleted
EC 3.4.21.41 complement subcomponent C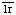
EC 3.4.21.42 complement subcomponent C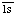
EC 3.4.21.43 classical-complement-pathway C3/C5 convertase
EC 3.4.21.44 deleted, included in EC 3.4.21.43
EC 3.4.21.45 complement factor I
EC 3.4.21.46 complement factor D
EC 3.4.21.47 alternative-complement-pathway C3/C5 convertase
EC 3.4.21.48 cerevisin
EC 3.4.21.49 hypodermin C
EC 3.4.21.50 lysyl endopeptidase
See the following file for:
EC 3.4.21.51 - EC 3.4.21.100 and EC 3.4.21.101 - EC 3.4.21.122
Entries
EC 3.4.21.1
Accepted name: chymotrypsin
Reaction: Preferential cleavage: Tyr , Trp
, Trp , Phe
, Phe , Leu
, Leu
Other names: chymotrypsins A and B; α-chymar ophth; avazyme; chymar; chymotest; enzeon; quimar; quimotrase; α-chymar; α-chymotrypsin A; α-chymotrypsin
Comments: Chymotrypsin A is formed from cattle and pig chymotrypsinogen A, several iso-forms being produced according to the number of bonds hydrolysed in the precursor. Formerly EC 3.4.4.5. Chymotrypsin B, formerly EC 3.4.4.6, formed from chymotrypsinogen B, is homologous with chymotrypsin A. Enzymes with specificity similar to that of chymotrypsins A and B have been isolated from many species. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
GTD,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9004-07-3
References
1. Wilcox, P.E. Chymotrypsinogens - chymotrypsins. Methods Enzymol. 19 (1970) 64-108
2. Blow, D.M. Structure and mechanism of chymotrypsin. Acc. Chem. Res. 9 (1976) 145-152
3. Bauer, C.-A. Active centers of α-chymotrypsin and of Streptomyces griseus proteases 1 and 3. Eur. J. Biochem. 105 (1980) 565-570. [PMID: 6768556]
4. Polgár, L. Structure and function of serine proteases. In New Comprehensive Biochemistry Vol. 16, Hydrolytic Enzymes (Neuberger, A. and Brocklehurst, K. eds), pp. 159-200 (1987) Elsevier, Amsterdam
5. Tomita, N., Izumoto, Y., Horii, A., Doi, S., Yokouchi, H., Ogawa, M., Mori, T. and Matsubara, K. Molecular cloning and nucleotide sequence of human pancreatic prechymotrypsinogen cDNA. Biochem. Biophys. Res. Commun. 158 (1989) 569-575. [PMID: 2917002]
[EC 3.4.21.1 created 1961 as EC 3.4.4.5 and EC 3.4.4.6, transferred 1972 to EC 3.4.21.1]
EC 3.4.21.2
Accepted name: chymotrypsin C
Reaction: Preferential cleavage: Leu , Tyr
, Tyr , Phe
, Phe , Met
, Met , Trp
, Trp , Gln
, Gln , Asn
, Asn
Comments: Formed from pig chymotrypsinogen C, and from cattle subunit II of procarboxypeptidase A. Reacts more readily with Tos-Leu-CH2Cl than Tos-Phe-CH2Cl in contrast to chymotrypsin. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9036-09-3
References
1. Peanasky, R.J., Gratecos, D., Baratti, J. and Rovery, M. Mode of activation and N-terminal sequence of subunit II in bovine procarboxypeptidase A and of porcine chymotrypsinogen C. Biochim. Biophys. Acta 181 (1969) 82-92. [PMID: 5792601]
2. Folk, J.E. Chymotrypsin C (porcine pancreas). Methods Enzymol. 19 (1970) 109-112
3. Wilcox, P.E. Chymotrypsinogens - chymotrypsins. Methods Enzymol. 19 (1970) 64-108
[EC 3.4.21.2 created 1972]
EC 3.4.21.3
Accepted name: metridin
Reaction: Preferential cleavage: Tyr , Phe
, Phe , Leu
, Leu ; little action on Trp-
; little action on Trp-
Other names: Metridium proteinase A; sea anemone protease A; sea anemone proteinase A
Comments: Digestive enzyme from the sea anemone Metridium senile.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 37288-75-8
References
1. Gibson, D. and Dixon, G.H. Chymotrypsin-like proteases from the sea anemone, Metridium senile. Nature 222 (1969) 753-756. [PMID: 4389140]
2. Stevenson, K.J., Gibson, D. and Dixon, G.H. Amino acid analyses of chymotrysin-like proteases from the sea anemone (Metridium senile). Can. J. Biochem. 52 (1974) 93-100. [PMID: 4150616]
[EC 3.4.21.3 created 1972]
EC 3.4.21.4
Accepted name: trypsin
Reaction: Preferential cleavage: Arg , Lys
, Lys
Other name(s): α-trypsin; β-trypsin; cocoonase; parenzyme; parenzymol; tryptar; trypure; pseudotrypsin; tryptase; tripcellim; sperm receptor hydrolase
Comments: The single polypeptide chain cattle β-trypsin is formed from trypsinogen by cleavage of one peptide bond. Further peptide bond cleavages produce α and other iso-forms. Isolated as multiple cationic and anionic trypsins [5] from the pancreas of many vertebrates and from lower species including crayfish, insects (cocoonase) and microorganisms (Streptomyces griseus) [3]. Type example of peptidase family S1. Formerly EC 3.4.4.4
Links to other databases:
BRENDA,
EXPASY,
KEGG,
GTD,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9002-07-7
References
1. Huber, R. and Bode, W. Structural basis of the activation and action of trypsin. Acc. Chem. Res. 11 (1978) 114-122
2. Walsh, K.A. Trypsinogens and trypsins of various species. Methods Enzymol. 19 (1970) 41-63
3. Read, R.J., Brayer, G.D., Jurásek, L. and James, M.N.G. Critical evaluation of comparative model building of Streptomyces griseus trypsin. Biochemistry 23 (1984) 6570-6575. [PMID: 6442164]
4. Fiedler, F. Effects of secondary interactions on the kinetics of peptide and peptide ester hydrolysis by tissue kallikrein and trypsin. Eur. J. Biochem. 163 (1987) 303-312. [PMID: 3643848]
5. Fletcher, T.S., Alhadeff, M., Craik, C.S. and Largman, C. Isolation and characterization of a cDNA encoding rat cationic trypsinogen. Biochemistry 26 (1987) 3081-3086. [PMID: 3112218]
6. Polgár, L. Structure and function of serine proteases. In New Comprehensive Biochemistry Vol. 16, Hydrolytic Enzymes (Neuberger, A. and Brocklehurst, K. eds), pp. 159-200 (1987) Elsevier, Amsterdam
7. Tani, T., Kawashima, I., Mita, K. and Takiguchi, Y. Nucleotide sequence of the human pancreatic trypsinogen III cDNA. Nucleic Acids Res. 18 (1990) 1631 only. [PMID: 2326201]
[EC 3.4.21.4 created 1961 as EC 3.4.4.4, transferred 1972 to EC 3.4.21.4]
EC 3.4.21.5
Accepted name: thrombin
Reaction: Selective cleavage of Arg Gly bonds in fibrinogen to form fibrin and release fibrinopeptides A and B
Gly bonds in fibrinogen to form fibrin and release fibrinopeptides A and B
Other names: fibrinogenase; thrombase; thrombofort; topical; thrombin-C; tropostasin; activated blood-coagulation factor II; blood-coagulation factor IIa; factor IIa; E thrombin; β-thrombin; γ-thrombin
Comments: Formed from prothrombin. More selective than trypsin and plasmin. In peptidase family S1 (trypsin family). Formerly EC 3.4.4.13
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9002-04-4
References
1. Baughman, D.J. Thrombin assay. Methods Enzymol. 19 (1970) 145-157
2. Magnusson, S. Bovine prothrombin and thrombin. Methods Enzymol. 19 (1970) 157-184
3. Miller, K.D. Horse prothrombin. Methods Enzymol. 19 (1970) 140-145
4. Lundblad, R.L., Kingdon, H.S. and Mann, K.G. Thrombin. Methods Enzymol. 45 (1976) 156-176. [PMID: 1011989]
5. Mann, K.G. Prothrombin. Methods Enzymol. 45 (1976) 123-156. [PMID: 1011988]
6. Davie, E.W., Fujikawa, K., Kurachi, K. and Kisiel, W. The role of serine proteases in the blood coagulation cascade. Adv. Enzymol. 48 (1979) 277-318. [PMID: 367103]
7. Cho, K., Tanaka, T., Cook, R.R., Kisiel, W., Fujikawa, K., Kurachi, K. and Powers, J.C. Active-site mapping of bovine and human blood coagulation serine proteases using synthetic peptide 4-nitroanilide and thio ester substrates. Biochemistry 23 (1984) 644-650. [PMID: 6370301]
8. MacGillivray, R.T.A. and Davie, E.W. Characterization of bovine prothrombin mRNA and its translation product. Biochemistry 23 (1984) 1626-1634. [PMID: 6326805]
[EC 3.4.21.5 created 1961 as EC 3.4.4.13, transferred 1972 to EC 3.4.21.5]
EC 3.4.21.6
Accepted name: coagulation factor Xa
Reaction: Selective cleavage of Arg Thr and then Arg
Thr and then Arg Ile bonds in prothrombin to form thrombin
Ile bonds in prothrombin to form thrombin
Other name(s): thrombokinase; prothrombase; prothrombinase; activated blood-coagulation factor X; autoprothrombin C; thromboplastin; plasma thromboplastin; factor Xa; activated Stuart-Prower factor; activated factor X
Comments: A blood coagulation factor formed from the proenzyme factor X by limited proteolysis.
Factor X is a glycoprotein composed of a heavy chain and a light chain, which are generated from a precursor protein by the excision of the tripeptide RKR and held together by one or more disulfide bonds. The activated factor Xa converts prothrombin to thrombin in the presence of factor Va, Ca2+ and phospholipids.
Scutelarin (EC 3.4.21.60) has similar specificity, but does not require factor Va.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9002-05-5
References:
1. Fujikawa, K. and Davie, E.W. Bovine factor X (Stuart factor). Methods Enzymol. 45 (1976) 89-95. [PMID: 1012041]
2. Jesty, J. and Nemerson, Y. The activation of bovine coagulation factor X. Methods Enzymol. 45 (1976) 95-107. [PMID: 1012042]
3. Davie, E.W., Fujikawa, K., Kurachi, K. and Kisiel, W. The role of serine proteases in the blood coagulation cascade. Adv. Enzymol. 48 (1979) 277-318. [PMID: 367103]
4. Jackson, C.M. and Nemerson, Y. Blood coagulation. Annu. Rev. Biochem. 49 (1980) 765-811. [PMID: 6996572]
5. McMullen, B.A., Fujikawa, K., Kisiel, W., Sasagawa, T., Howald, W.N., Kwa, E.Y. and Weinstein, B. Complete amino acid sequence of the light chain of human blood coagulation factor X: evidence for identification of residue 63 as β-hydroxyaspartic acid. Biochemistry 22 (1983) 2875-2884. [PMID: 6871167]
6. Cho, K., Tanaka, T., Cook, R.R., Kisiel, W., Fujikawa, K., Kurachi, K. and Powers, J.C. Active-site mapping of bovine and human blood coagulation serine proteases using synthetic peptide 4-nitroanilide and thio ester substrates. Biochemistry 23 (1984) 644-650. [PMID: 6370301]
[EC 3.4.21.6 created 1972, modified 2011]
EC 3.4.21.7
Accepted name: plasmin
Reaction: Preferential cleavage: Lys > Arg
> Arg ; higher selectivity than trypsin. Converts fibrin into soluble products
; higher selectivity than trypsin. Converts fibrin into soluble products
Other names: fibrinase; fibrinolysin; actase; serum tryptase; thrombolysin
Comments: Formed from plasminogen by proteolysis which results in multiple forms of the active plasmin. In peptidase family S1 (trypsin family). Formerly EC 3.4.4.14
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9001-90-5
References
1. Castellino, F.J. and Sodetz, J.M. Rabbit plasminogen and plasmin isozymes. Methods Enzymol. 45 (1976) 273-286. [PMID: 138065]
2. Castellino, F.J. and Powell, J.R. Human plasminogen. Methods Enzymol. 80 (1981) 365-378. [PMID: 6210827]
3. Robbins, K.C., Summaria, L. and Wohl, R.C. Human plasmin. Methods Enzymol. 80 (1981) 379-387. [PMID: 6210828]
[EC 3.4.21.7 created 1961 as EC 3.4.4.14, transferred 1972 to EC 3.4.21.7]
[EC 3.4.21.8 Transferred entry: now EC 3.4.21.34 - Plasma kallikrein and EC 3.4.21.35 - Tissue kallikrein (EC 3.4.21.8 created 1972, deleted 1981)]
EC 3.4.21.9
Accepted name: enteropeptidase
Reaction: Activation of trypsinogen by selective cleavage of Lys6 Ile bond
Ile bond
Other names: enterokinase
Comments: Is not inhibited by protein inhibitors of trypsin. In peptidase family S1 (trypsin family). Formerly EC 3.4.4.8
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9014-74-8
References
1. Light, A. and Janska, H. Enterokinase (enteropeptidase): comparative aspects. Trends Biochem. Sci. 14 (1989) 110-112. [PMID: 2658218]
[EC 3.4.21.9 created 1961 as EC 3.4.4.8, transferred 1972 to EC 3.4.21.9]
EC 3.4.21.10
Accepted name: acrosin
Reaction: Preferential cleavage: Arg , Lys
, Lys
Other name(s): acrosomal proteinase; acrozonase; α-acrosin; β-acrosin; ψ-acrosin; acrosomal protease; acrosin amidase
Comments: Occurs in spermatozoa; formed from proacrosin by limited proteolysis. Inhibited by naturally occurring trypsin inhibitors. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9068-57-9
References
1. Müller-Esterl, W. and Fritz, H. Sperm acrosin. Methods Enzymol. 80 (1981) 621-632. [PMID: 7043204]
2. Skoog, M.T., Mehdi, S., Wiseman, J.S. and Bey, P. The specificity of two proteinases that cleave adjacent to arginine, C1 esterase and acrosin, for peptide p-nitroanilide substrates. Biochim. Biophys. Acta 996 (1989) 89-94. [PMID: 2500154]
3. Keime, S., Adham, I.M. and Engel, W. Nucleotide sequence and exon-intron organisation of the human proacrosin gene. Eur. J. Biochem. 190 (1990) 195-200. [PMID: 2114285]
[EC 3.4.21.10 created 1972]
[EC 3.4.21.11 Transferred entry: now EC 3.4.21.36 - pancreatic elastase and EC 3.4.21.37 - leukocyte elastase (EC 3.4.21.11 created 1972, deleted 1981)]
EC 3.4.21.12
Accepted name: α-lytic endopeptidase
Reaction: Preferential cleavage: Ala , Val
, Val in bacterial cell walls, elastin and other proteins
in bacterial cell walls, elastin and other proteins
Other names: myxobacter α-lytic proteinase; α-lytic proteinase; α-lytic protease; Mycobacterium sorangium α-lytic proteinase; Myxobacter 495 α-lytic proteinase; α-lytic proteinase; Myxobacter α-lytic proteinase; Mycobacterium sorangium α-lytic proteinase
Comments: From the myxobacterium Lysobacter enzymogenes. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 37288-76-9
References
1. Olson, M.O.J., Nagabushan, N., Dzwiniel, M., Smillie, L.B. and Whitaker, D.R. Primary structure of α-lytic protease: a bacterial homologue of the pancreatic serine proteases. Nature 228 (1970) 438-442. [PMID: 5482494]
2. Polgár, L. Structure and function of serine proteases. In New Comprehensive Biochemistry Vol. 16, Hydrolytic Enzymes (Neuberger, A. and Brocklehurst, K. eds), pp. 159-200 (1987) Elsevier, Amsterdam
3. Epstein, D.M. and Wensink, P.C. The α-lytic protease gene of Lysobacter enzymogenes. The nucleotide sequence predicts a large prepro-peptide with homology to pro-peptides of other chymotrypsin-like enzymes. J. Biol. Chem. 263 (1988) 16586-16590. [PMID: 3053694]
4. Bone, R., Frank, D., Kettner, C.A. and Agard, D.A. Structural analysis of specificity: α-lytic protease complexes with analogues of reaction intermediates. Biochemistry 28 (1989) 7600-7609. [PMID: 2611204]
[EC 3.4.21.12 created 1972]
[EC 3.4.21.13 Transferred entry: now EC 3.4.16.5, carboxypeptidase C and EC 3.4.16.6, carboxypeptidase D(EC 3.4.21.13 created 1972, deleted 1978)]
[EC 3.4.21.14 Transferred entry: now EC 3.4.21.62 - subtilisin; EC 3.4.21.63 - oryzin; EC 3.4.21.64 - endopeptidase K; EC 3.4.21.65 - thermomycolin; and EC 3.4.21.67 - endopeptidase So (EC 3.4.21.14 created 1961 as EC 3.4.4.16, transferred 1972 to EC 3.4.21.14, modified 1986, deleted 1992)]
[EC 3.4.21.15 Transferred entry: now EC 3.4.21.63 - oryzin (EC 3.4.21.15 created 1972, deleted 1978 [transferred to EC 3.4.21.14, deleted 1992])]
[EC 3.4.21.16 Deleted entry: Alternaria serine proteinase (EC 3.4.21.16 created 1972, deleted 1992)]
[EC 3.4.21.17 Deleted entry: Arthrobacter serine proteinase (EC 3.4.21.17 created 1972, deleted 1978 [transferred to EC 3.4.21.14, deleted 1992])]
[EC 3.4.21.18 Deleted entry: Tenebrio α-proteinase (EC 3.4.21.18 created 1972 [EC 3.4.99.24 created 1972, incorporated 1978], deleted 1992)]
EC 3.4.21.19
Accepted name: glutamyl endopeptidase
Reaction: Preferential cleavage: Glu , Asp
, Asp
Other names: V8 proteinase; endoproteinase Glu-C; staphylococcal serine proteinase
Comments: From Staphylococcus aureus strain V8. In appropriate buffer the specificity is restricted to Glu . In peptidase family S1 (trypsin family)
. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 137010-42-5
References
1. Drapeau, G.R. Protease from Staphylococcus aureus. Methods Enzymol. 45 (1976) 469-475. [PMID: 1012010]
2. Drapeau, G.R. The primary structure of staphylococcal protease. Can. J. Biochem. 56 (1978) 534-544. [PMID: 96922]
3. Carmona C. and Gray G.L. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res.15 (1987) 6757 only. [PMID: 3306605]
[EC 3.4.21.19 created 1978]
EC 3.4.21.20
Accepted name: cathepsin G
Reaction: Specificity similar to chymotrypsin C
Other name(s): chymotrypsin-like proteinase; neutral proteinase
Comments: From azurophil granules of polymorphonuclear leukocytes. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 56645-49-9
References
1. Barrett, A.J. Cathepsin G. Methods Enzymol. 80 (1981) 561-565. [PMID: 7341917]
2. Tanaka, T., Minematsu, Y., Reilly, C.F., Travis, J. and Powers, J.C. Human leukocyte cathepsin G. Subsite mapping with 4-nitroanilides, chemical modification, and effect of possible cofactors. Biochemistry 24 (1985) 2040-2047. [PMID: 4016099]
3. Hohn, P.A., Popescu, N.C., Hanson, R.D., Salvesen, G. and Ley, T.J. Genomic organization and chromosomal localization of the human cathepsin G gene. J. Biol. Chem. 264 (1989) 13412-13419. [PMID: 2569462]
[EC 3.4.21.20 created 1978]
EC 3.4.21.21
Accepted name: coagulation factor VIIa
Reaction: Selective cleavage of Arg Ile bond in factor X to form factor Xa
Ile bond in factor X to form factor Xa
Other name(s): blood-coagulation factor VIIa; activated blood coagulation factor VII
Comments: Formed from the precursor factor VII. The cattle enzyme is more readily inhibited by diisopropyl fluorophosphate than the human [1]. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 65312-43-8
References
1. Nemerson, Y. and Esnouf, M.P. Activation of a proteolytic system by a membrane lipoprotein: mechanism of action of tissue factor. Proc. Natl. Acad. Sci. USA 70 (1973) 310-314. [PMID: 4510277]
2. Davie, E.W., Fujikawa, K., Kurachi, K. and Kisiel, W. The role of serine proteases in the blood coagulation cascade. Adv. Enzymol. 48 (1979) 277-318. [PMID: 367103]
3. Jackson, C.M. and Nemerson, Y. Blood coagulation. Annu. Rev. Biochem. 49 (1980) 765-811. [PMID: 6996572]
4. Broze, G.J., Jr and Majerus, P.W. Human factor VII. Methods Enzymol. 80 (1981) 228-237
[EC 3.4.21.21 created 1978]
EC 3.4.21.22
Accepted name: coagulation factor IXa
Reaction: Selective cleavage of Arg Ile bond in factor X to form factor Xa
Ile bond in factor X to form factor Xa
Other names: activated Christmas factor; blood-coagulation factor IXa; activated blood-coagulation factor IX; autoprothrombin II; blood platelet cofactor II; activated blood coagulation factor XI
Comments: A chymotrypsin homologue, and one of the γ-carboxyglutamic acid-containing blood coagulation factors. The proenzyme factor IX is activated by factor XIa. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 37316-87-3
References
1. Fujikawa, K. and Davie, E.W. Bovine factor IX (Christmas factor). Methods Enzymol. 45 (1976) 74-83. [PMID: 1012029]
2. Davie, E.W., Fujikawa, K., Kurachi, K. and Kisiel, W. The role of serine proteases in the blood coagulation cascade. Adv. Enzymol. 48 (1979) 277-318. [PMID: 367103]
3. Link, R.P. and Castellino, F.J. Kinetic comparison of bovine blood coagulation factor IXaα and IXaβ towards bovine factor X. Biochemistry 22 (1983) 4033-4041. [PMID: 6412750]
4. Cho, K., Tanaka, T., Cook, R.R., Kisiel, W., Fujikawa, K., Kurachi, K. and Powers, J.C. Active-site mapping of bovine and human blood coagulation serine proteases using synthetic peptide 4-nitroanilide and thio ester substrates. Biochemistry 23 (1984) 644-650. [PMID: 6370301]
[EC 3.4.21.22 created 1978]
[EC 3.4.21.23 Deleted entry: Vipera russelli proteinase (EC 3.4.21.23 created 1978, deleted 1992)]
[EC 3.4.21.24 Deleted entry: red cell neutral endopeptidase (EC 3.4.21.24 created 1978, deleted 1992)]
EC 3.4.21.25
Accepted name: cucumisin
Reaction: Hydrolysis of proteins with broad specificity
Other name(s): euphorbain; solanain; hurain; tabernamontanain
Comments: From the sarcocarp of the musk melon (Cucumis melo). In peptidase family S8 (subtilisin family). Other endopeptidases from plants, which are less well characterized but presumably of serine-type, include euphorbain from Euphorbia cerifera [6], solanain from horse-nettle Solanum elaeagnifolium [1], hurain from Hura crepitans [2] and tabernamontanain from Tabernamontana grandiflora [3].
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 82062-89-3
References
1. Greenberg, D.M. and Winnick, T. Plant proteases. I. Activation-inhibition reactions. J. Biol. Chem. 135 (1940) 761-773
2. Jaffé, W.G. Hurain, a new plant protease from Hura crepitans. J. Biol. Chem. 149 (1943) 1-7
3. Jaffé, W.G. A new vegetable proteolytic enzyme of the papain class. Rev. Brasil Biol. 3 (1943) 149-157
4. Kaneda, M. and Toninaga, N. Isolation and characterization of a proteinase from the sarcocarp of melon fruit. J. Biochem. (Tokyo) 78 (1975) 1287-1296. [PMID: 5423]
5. Kaneda, M., Ohmine, H., Yonezawa, H. and Tominaga, N. Amino acid sequence around the reactive site of cucumisin from melon fruit. J. Biochem. (Tokyo) 95 (1984) 825-829. [PMID: 6427203]
6. Lynn, K.R. and Clevette-Radford, N.A. Two proteases from the latex of Elaeophorbia drupifera. Phytochemistry 24 (1985) 2843-2845
7. Kaneda, N., Minematsu, Y., Powers, J.C. and Tominaga, N. Specificity of cucumisin in hydrolysis of peptide thiobenzyl esters. Agric. Biol. Chem. 50 (1986) 1075-1076
[EC 3.4.21.25 created 1978 (EC 3.4.21.56 created 1972 as EC 3.4.99.7 transferred 1989 to EC 3.4.21.56, deleted 1992; EC 3.4.99.9 created 1972 deleted 1992; EC 3.4.99.21 created 1972 deleted 1992; EC 3.4.99.23 created 1972 deleted 1992; all covered by EC 3.4.21.25)]
EC 3.4.21.26
Accepted name: prolyl oligopeptidase
Reaction: Hydrolysis of —Pro and to a lesser extent —Ala
and to a lesser extent —Ala in oligopeptides
in oligopeptides
Other names: post-proline cleaving enzyme; proline-specific endopeptidase; post-proline endopeptidase; proline endopeptidase; endoprolylpeptidase; prolyl endopeptidase
Comments: Found in vertebrates, plants and Flavobacterium. Generally cytosolic, commonly activated by thiol compounds. Type example of peptidase family S9
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 72162-84-6
References
1. Walter, R. and Yoshimoto, T. Postproline cleaving enzyme: kinetic studies of size and stereospecificity of its active site. Biochemistry 17 (1978) 4139-4144. [PMID: 708698]
2. Nomura, K. Specificity of prolyl endopeptidase. FEBS Lett. 209 (1986) 235-237. [PMID: 3539636]
3. Moriyama, A., Nakanishi, M. and Sasaki, M. Porcine muscle prolyl endopeptidase and its endogenous substrates. J. Biochem. (Tokyo) 104 (1988) 112-117. [PMID: 2851585]
4. Rennex, D., Hemmings, B.A., Hofsteenge, J. and Stone, S.R. cDNA cloning of porcine brain prolyl endopeptidase and identification of the active-site seryl residue. Biochemistry, 30 (1991) 2195-2203. [PMID: 1900195]
[EC 3.4.21.26 created 1978, modified 1981 (EC 3.4.22.18 created 1981, incorporated 1992)]
EC 3.4.21.27
Accepted name: coagulation factor XIa
Reaction: Selective cleavage of Arg Ala and Arg
Ala and Arg Val bonds in factor IX to form factor IXa
Val bonds in factor IX to form factor IXa
Other name(s): blood-coagulation factor XIa; activated blood-coagulation factor XI; activated plasma thromboplastin antecedent
Comments: In peptidase family S1 (trypsin family), and one of the γ-carboxyglutamic acid-containing blood coagulation factors. The proenzyme factor XI is activated by factor XIIa
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 37203-61-5
References
1. Kurachi, K. and Davie, E.W. Human factor XI (plasma thromboplastin antecedent). Methods Enzymol. 80 (1981) 211-220
2. Cho, K., Tanaka, T., Cook, R.R., Kisiel, W., Fujikawa, K., Kurachi, K. and Powers, J.C. Active-site mapping of bovine and human blood coagulation serine proteases using synthetic peptide 4-nitroanilide and thio ester substrates. Biochemistry 23 (1984) 644-650. [PMID: 6370301]
3. Fujikawa, K., Chung, D.W., Hendrickson, L.E. and Davie, E.W. Amino acid sequence of human factor XI, a blood coagulation factor with four tandem repeats that are highly homologous with plasma prekallikrein. Biochemistry 25 (1986) 2417-2424. [PMID: 3636155]
[EC 3.4.21.27 created 1978]
[EC 3.4.21.28 Transferred entry: now included with EC 3.4.21.74 - venombin A (EC 3.4.21.28 created 1978, deleted 1992)]
[EC 3.4.21.29 Transferred entry: now included with EC 3.4.21.74 - venombin A (EC 3.4.21.29 created 1978, deleted 1992)]
[EC 3.4.21.30 Transferred entry: now included with EC 3.4.21.74 - venombin A (EC 3.4.21.30 created 1978, deleted 1992)]
[EC 3.4.21.31 Transferred entry: now EC 3.4.21.68 - t-plasminogen activator and EC 3.4.21.73 - u-plasminogen activator (EC 3.4.21.31 created 1972 as EC 3.4.99.26, transferred 1978 to EC 3.4.21.31, deleted 1992)]
EC 3.4.21.32
Accepted name: brachyurin
Reaction: Hydrolysis of proteins, with broad specificity for peptide bonds. Native collagen is cleaved about 75% of the length of the molecule from the N-terminus. Low activity on small molecule substrates of both trypsin and chymotrypsin
Other names: Uca pugilator collagenolytic proteinase; crab protease I; crab protease II
Comments: From hepatopancreas of the fiddler crab, Uca pugilator. In peptidase family S1 (trypsin family). Other serine endopeptidases that degrade collagen, but are not listed separately here, include a second endopeptidases from Uca pugilator [4], digestive enzymes from other decapod crustacea [5,6], and an enzyme from the fungus Entomophthora coronata (formerly EC 3.4.21.33) [1]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 848900-32-3
References
1. Hurion, N., Fromentin, H. and Keil, B. Specificity of the collagenolytic enzyme from the fungus Entomophthora coronata: comparison with the bacterial collagenase from Achromobacter iophagus. Arch. Biochem. Biophys. 192 (1979) 438-445. [PMID: 219780]
2. Grant, G.A., Eisen, A.Z. and Bradshaw, R.A. Collagenolytic protease from fiddler crab (Uca pugilator). Methods Enzymol. 80 (1981) 722-734
3. Welgus, H.G., Grant, G.A., Jeffrey, J.J. and Eisen, A.Z. Substrate specificity of the collagenolytic serine protease from Uca pugilator: studies with collagenous substrates. Biochemistry 21 (1982) 5183-5189. [PMID: 6756469]
4. Welgus, H.G. and Grant, G.A. Degradation of collagen substrates by a trypsin-like serine protease form the fiddler crab Uca pugilator. Biochemistry 22 (1983) 2228-2233. [PMID: 6305411]
5. Klimova, O.A., Borukhov, S.I., Solovyeva, N.I., Balaevskaya, T.O. and Strongin, A.Y. The isolation and properties of collagenolytic proteases from crab hepatopancreas. Biochem. Biophys. Res. Commun. 166 (1990) 1411-1420. [PMID: 2154979]
6. Lu, P.-J., Liu, H.-C. and Tsai, I.-H. The midgut trypsins of shrimp (Penaeus monodon). High efficiency toward native protein substrates including collagens. Biol. Chem. Hoppe-Seyler 371 (1990) 851-859. [PMID: 1963309]
[EC 3.4.21.32 created 1978]
[EC 3.4.21.33 Deleted entry: Entomophthora collagenolytic proteinase (EC 3.4.21.33 created 1978, deleted 1992)]
EC 3.4.21.34
Accepted name: plasma kallikrein
Reaction: Selective cleavage of some Arg and Lys
and Lys bonds, including Lys
bonds, including Lys Arg and Arg
Arg and Arg Ser in (human) kininogen to release bradykinin
Ser in (human) kininogen to release bradykinin
Other names: serum kallikrein; kininogenin; kallikrein I; kallikrein II; kininogenase; kallikrein; callicrein; glumorin; padreatin; padutin; kallidinogenase; bradykininogenase; panceatic kallikrein; onokrein P; dilminal D; depot-Padutin; urokallikrein; urinary kallikrein
Comments: Formed from plasma prokallikrein (Fletcher factor) by factor XIIa. Activates coagulation factors XII, VII and plasminogen. Selective for Arg > Lys in P1, in small molecule substrates. In peptidase family S1 (trypsin family). Formerly EC 3.4.4.21 and included in EC 3.4.21.8
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 410538-33-9
References
1. Heimark, R.L. and Davie, E.W. Bovine and human plasma prekallikrein. Methods Enzymol. 80 (1981) 157-172. [PMID: 6918767]
2. McRae, B.J., Kurachi, K., Heimark, R.L., Fujikawa, K., Davie, E.W. and Powers, J.C. Mapping the active sites of bovine thrombin, factor IXa, factor Xa, factor XIa, factor XIIa, plasma kallikrein, and trypsin with amino acid and peptide thioesters: development of new sensitive substrates. Biochemistry 20 (1981) 7196-7206. [PMID: 6976185]
3. Silverberg, M. and Kaplan, A.P. Prekallikrein. Methods Enzymol. 163 (1988) 85-95. [PMID: 3237096]
4. Seidah, N.G., Ladenheim, R., Mbikay, M., Hamelin, J., Lutfalla, G., Rougeon, F., Lazure, C. and Chrétien, M. The cDNA structure of rat plasma kallikrein. DNA 8 (1989) 563-574. [PMID: 2598771]
5. Tsuda, Y., Teno, N., Okada, Y., Wanaka, K., Bohgaki, M., Hijikata-Okunomiya, A., Okamoto, U., Naito, T. and Okamoto, S. Synthesis of tripeptide chloromethyl ketones and examination of their inhibitory effects on plasmin and plasma kallikrein. Chem. Pharm. Bull. 37 (1989) 3108-3111. [PMID: 2534361]
[EC 3.4.21.34 created 1965 as EC 3.4.4.21, transferred 1972 to EC 3.4.21.8, part transferred 1981 to EC 3.4.21.34]
EC 3.4.21.35
Accepted name: tissue kallikrein
Reaction: Preferential cleavage of Arg bonds in small molecule substrates. Highly selective action to release kallidin (lysyl-bradykinin) from kininogen involves hydrolysis of Met
bonds in small molecule substrates. Highly selective action to release kallidin (lysyl-bradykinin) from kininogen involves hydrolysis of Met or Leu
or Leu . The rat enzyme is unusual in liberating bradykinin directly from autologous kininogens by cleavage at two Arg
. The rat enzyme is unusual in liberating bradykinin directly from autologous kininogens by cleavage at two Arg bonds [5]
bonds [5]
Other names: glandular kallikrein; pancreatic kallikrein; submandibular kallikrein; submaxillary kallikrein; kidney kallikrein; urinary kallikrein; kallikrein; salivary kallikrein; kininogenin; kininogenase; callicrein; glumorin; padreatin; padutin; kallidinogenase; bradykininogenase; depot-padutin; urokallikrein; dilminal D; onokrein P
Comments: Formed from tissue prokallikrein by activation with trypsin. In peptidase family S1 (trypsin family). Formerly EC 3.4.4.21 and included in EC 3.4.21.8. A large number of tissue kallikrein-related sequences have been reported for rats [16] and mice [7], though fewer seem to exist in other mammals. The few that have been isolated and tested on substrates include mouse γ-renin (EC 3.4.21.54), submandibular proteinase A (formerly EC 3.4.21.40), [2,15], epidermal growth-factor-binding protein, nerve growth factor γ-subunit, rat tonin [3,4,9], submaxillary proteinases A and B [10], T-kininogenase [18], kallikreins k7 and k8 [17] and human prostate-specific antigen (γ-seminoprotein, [6])
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 389069-73-2
References
1. Fiedler, F., Fink, E., Tschesche, H. and Fritz, H. Porcine glandular kallikreins. Methods Enzymol. 80 (1981) 493-532. [PMID: 7043199]
2. Anundi, H., Ronne, H., Peterson, P.A. and Rask, L. Partial amino-acid sequence of the epidermal growth-factor-binding protein. Eur. J. Biochem. 129 (1982) 365-371. [PMID: 6295764]
3. Pesquero, J.L., Boschcov, P., Oliveira, M.C.F. and Paiva, A.C.M. Effect of substrate size on tonin activity. Biochem. Biophys. Res. Commun. 108 (1982) 1441-1446. [PMID: 6295383]
4. Gutkowska, J., Corvol, P., Figueiredo, A.F.S., Inagami, T., Bouhnik, J. and Genest, J. Kinetic studies of rat renin and tonin on purified rat angiotensinogen. Can. J. Biochem. Cell Biol. 62 (1984) 136-142. [PMID: 6043136]
5. Kato, H., Enjyoji, K., Miyata, T., Hayashi, I., Oh-Ishi, S. and Iwanaga, S. Demonstration of arginyl-bradykinin moiety in rat HMW kininogen: direct evidence for liberation of bradykinin by rat glandular kallikreins. Biochem. Biophys. Res. Commun. 127 (1985) 289-295. [PMID: 3844939]
6. Akiyama, K., Nakamura, T., Iwanaga, S. and Hara, M. The chymotrypsin-like activity of human prostate-specific antigen, γ-seminoprotein. FEBS Lett. 225 (1987) 168-172. [PMID: 3691800]
7. Evans, B.A., Drinkwater, C.C. and Richards, R.I. Mouse glandular kallikrein genes. Structure and partial sequence analysis of the kallikrein gene locus. J. Biol. Chem. 262 (1987) 8027-8034. [PMID: 3036794]
8. Fiedler, F. Effects of secondary interactions on the kinetics of peptide and peptide ester hydrolysis by tissue kallikrein and trypsin. Eur. J. Biochem. 163 (1987) 303-312. [PMID: 7043199]
9. Fujinaga, M. and James, M.N.G. Rat submaxillary gland serine protease, tonin. Structure solution and refinement at 1.8 Å resolution. J. Mol. Biol. 195 (1987) 373-396. [PMID: 2821276]
10. Kato, H., Nakanishi, E., Enjyoji, K., Hayashi, I., Oh-ishi, S. and Iwanaga, S. Characterization of serine proteinases isolated from rat submaxillary gland: with special reference to the degradation of rat kininogens by these enzymes. J. Biochem. (Tokyo) 102 (1987) 1389-1404. [PMID: 3844939]
11. Bailey, G.S. Rat pancreas kallikrein. Methods Enzymol. 163 (1989) 115-128. [PMID: 3237072]
12. Blaber, M., Isackson, P.J., Marsters, J.C., Jr, Burnier, J.P. and Bradshaw, R.A. Substrate specificities of growth factor associated kallikreins of the mouse submandibular gland. Biochemistry 28 (1988) 7813-7819. [PMID: 2611215]
13. Chao, J. and Chao, L. Rat urinary kallikrein. Methods Enzymol. 163 (1988) 128-143. [PMID: 3070295]
14. Geiger, R. and Miska, W. Human tissue kallikrein. Methods Enzymol. 163 (1988) 102-115. [PMID: 2975076]
15. Bertrand, R., Derancourt, J. and Kassab, R. Selective cleavage at lysine of the 50 kDa-20 kDa connector loop segment of skeletal myosin S-1 by endoproteinase Arg-C. FEBS Lett. 246 (1989) 171-176. [PMID: 2523317]
16. Wines, D.R., Brady, J.M., Pritchett, D.B., Roberts, J.L. and MacDonald, R.J. Organization and expression of the rat kallikrein gene family. J. Biol. Chem. 264 (1989) 7653-7662. [PMID: 2708383]
17. Elmoujahed, A., Gutman, N., Brillard, M. and Gauthier, F. Substrate specificity of two kallikrein family gene products isolated from the rat submaxillary gland. FEBS Lett. 265 (1990) 137-140. [PMID: 2194829]
18. Xiong, W., Chen, L.-M. and Chao, J. Purification and characterization of a kallikrein-like T-kininogenase. J. Biol. Chem. 265 (1990) 2822-2827. [PMID: 2303430]
[EC 3.4.21.35 created 1965 as EC 3.4.4.21, transferred 1972 to EC 3.4.21.8, part transferred 1981 to EC 3.4.21.35]
EC 3.4.21.36
Accepted name: pancreatic elastase
Reaction: Hydrolysis of proteins, including elastin. Preferential cleavage: Ala
Other names: pancreatopeptidase E; pancreatic elastase I; elastase; elaszym; serine elastase
Comments: Formed by activation of proelastase from mammalian pancreas by trypsin. In peptidase family S1 (trypsin family). Formerly included in EC 3.4.21.11
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9004-06-2
References
1. Shotton, D.M. Elastase. Methods Enzymol. 19 (1970) 113-140
2. Harper, J.W., Cook, R.R., Roberts, C.J., McLaughlin, B.J. and Powers, J.C. Active site mapping of the serine proteases human leukocyte elastase, cathepsin G, porcine pancreatic elastase, rat mast cell proteases I and II, bovine chymotrypsin Aα, and Staphylococcus aureus protease V-8 using tripeptide thiobenzyl ester substrates. Biochemistry 23 (1984) 2995-3002. [PMID: 6380580]
3. Kawashima, I., Tani, T., Shimoda, K. and Takiguchi, Y. Characterization of pancreatic elastase II cDNAs: two elastase II mRNAs are expressed in human pancreas. DNA 6 (1987) 163-172. [PMID: 3646943]
4. Bieth, J.G., Dirrig, S., Jung, M.-L., Boudier, C., Papamichael, E., Sakarellos, C. and Dimicoli, J.-L. Investigation of the active center of rat pancreatic elastase. Biochim. Biophys. Acta 994 (1989) 64-74. [PMID: 2909256]
5. Bode, W., Meyer, E., Jr and Powers, J.C. Human leukocyte and porcine pancreatic elastase: X-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry 28 (1989) 1951-1963 . [PMID: 2655701]
[EC 3.4.21.36 created 1981 (EC 3.4.4.7 created 1961, transferred 1972 to EC 3.4.21.11 created 1972, part incorporated 1984)]
EC 3.4.21.37
Accepted name: leukocyte elastase
Reaction: Hydrolysis of proteins, including elastin. Preferential cleavage Val > Ala
> Ala
Other names: lysosomal elastase; neutrophil elastase; polymorphonuclear leukocyte elastase; elastase; elaszym; serine elastase; lysosomal elastase; granulocyte elastase
Comments: Differs from pancreatic elastase in specificity on synthetic substrates and in inhibitor sensitivity. In peptidase family S1 (trypsin family). Formerly included in EC 3.4.21.11
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 9004-06-2
References
1. Barrett, A.J. Leukocyte elastase. Methods Enzymol. 80 (1981) 581-588. [PMID: 7043201]
2. Harper, J.W., Cook, R.R., Roberts, C.J., McLaughlin, B.J. and Powers, J.C. Active site mapping of the serine proteases human leukocyte elastase, cathepsin G, porcine pancreatic elastase, rat mast cell proteases I and II, bovine chymotrypsin Aα, and Staphylococcus aureus protease V-8 using tripeptide thiobenzyl ester substrates. Biochemistry 23 (1984) 2995-3002. [PMID: 6380580]
3. Stein, R.L., Strimpler, A.M., Hori, H. and Powers, J.C. Catalysis by human leukocyte elastase: mechanistic insights into specificity requirements. Biochemistry 26 (1987) 1301-1305. [PMID: 3646070]
4. Bode, W., Meyer, E., Jr and Powers, J.C. Human leukocyte and porcine pancreatic elastase: X-ray crystal structures, mechanism, substrate specificity, and mechanism-based inhibitors. Biochemistry 28 (1989) 1951-1963 . [PMID: 2655701]
[EC 3.4.21.37 created 1981 (EC 3.4.4.7 created 1961, transferred 1972 to EC 3.4.21.11 created 1972, part incorporated 1984)]
EC 3.4.21.38
Accepted name: coagulation factor XIIa
Reaction: Selective cleavage of Arg Ile bonds in factor VII to form factor VIIa and factor XI to form factor XIa
Ile bonds in factor VII to form factor VIIa and factor XI to form factor XIa
Other names: Hageman factor (activated); blood-coagulation factor XIIf; activated β blood-coagulation factor XII; prealbumin activator; Hageman factor β-fragment; Hageman factor fragment HFf; blood-coagulation factor XIIaβ; prekallikrein activator; kallikreinogen activator
Comments: Also activates plasminogen and plasma prokallikrein. Formed from the proenzyme, factor XII, by plasma kallikrein or factor XIIa. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 75216-42-1
References
1. Fujikawa, K. and Davie, E.W. Human factor XII (Hageman factor). Methods Enzymol. 80 (1981) 198-211. [PMID: 6918768]
2. Cho, K., Tanaka, T., Cook, R.R., Kisiel, W., Fujikawa, K., Kurachi, K. and Powers, J.C. Active-site mapping of bovine and human blood coagulation serine proteases using synthetic peptide 4-nitroanilide and thio ester substrates. Biochemistry 23 (1984) 644-650. [PMID: 6370301]
3. Que, B.G. and Davie, E.W. Characterization of a cDNA coding for human factor XII (Hageman factor). Biochemistry 25 (1986) 1525-1528 . [PMID: 3011063]
4. Fujikawa, K. Bovine Hageman factor and its fragments. Methods Enzymol. 163 (1988) 54-68. [PMID: 3237089]
5. Silverberg, M. and Kaplan, A.P. Human Hageman factor and its fragments. Methods Enzymol. 163 (1988) 68-80. [PMID: 3237092]
[EC 3.4.21.38 created 1981]
EC 3.4.21.39
Accepted name: chymase
Reaction: Preferential cleavage: Phe > Tyr
> Tyr > Trp
> Trp > Leu
> Leu
Other names: mast cell protease I; skeletal muscle protease; skin chymotryptic proteinase; mast cell serine proteinase; skeletal muscle (SK) protease
Comments: In mast cell granules. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 97501-92-3
References
1. Woodbury, R.G., Everitt, M. and Neurath, H. Mast cell proteases. Methods Enzymol. 80 (1981) 588-609. [PMID: 7043202]
2. Powers, J.C., Tanaka, T., Harper, J.W., Minematsu, Y., Barker, L., Lincoln, D., Crumley, K.V., Fraki, J.E., Schechter, N.M., Lazarus, G.G., Nakajima, K., Nakashino, K., Neurath, H. and Woodbury, R.G. Mammalian chymotrypsin-like enzymes. Comparative reactivities of rat mast cell proteases, human and dog skin chymases, and human cathepsin G with peptide 4-nitroanilide substrates and with peptide chloromethyl ketone and sulfonyl fluoride inhibitors. Biochemistry 24 (1985) 2048-2058. [PMID: 3893542]
3. Johnson, L.A., Moon, K.E. and Eisenberg, M. Purification to homogeneity of the human skin chymotryptic proteinase "chymase". Anal. Biochem. 155 (1986) 358-364. [PMID: 2425663]
[EC 3.4.21.39 created 1981]
[EC 3.4.21.40 Deleted entry: submandibular proteinase A (EC 3.4.21.40 created 1981, deleted 1992)]
EC 3.4.21.41
Accepted name: complement subcomponent C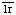
Reaction: Selective cleavage of Lys(or Arg) Ile bond in complement subcomponent C1s to form C
Ile bond in complement subcomponent C1s to form C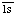 (EC 3.4.21.42)
(EC 3.4.21.42)
Other name(s): activated complement C1r; C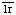 esterase; activated complement C1r
esterase; activated complement C1r
Comments: Activated from proenzyme C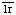 in plasma during activation of the complement system by the "classical" route. In peptidase family S1 (trypsin family)
in plasma during activation of the complement system by the "classical" route. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 80295-69-8
References
1. Sim, R.B. The human complement system serine proteases C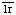 and C
and C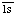 and their proenzymes. Methods Enzymol. 80 (1981) 26-42. [PMID: 6281620]
and their proenzymes. Methods Enzymol. 80 (1981) 26-42. [PMID: 6281620]
2. Leytus, S.P., Kurachi, K., Sakariassen, K.S. and Davie, E.W. Nucleotide sequence of the cDNA coding for human complement C1r. Biochemistry 25 (1986) 4855-4863. [PMID: 3021205]
3. Müller-Eberhard, H.J. Molecular organization and function of the complement system. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
[EC 3.4.21.41 created 1981]
EC 3.4.21.42
Accepted name: complement subcomponent C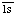
Reaction: Cleavage of Arg Ala bond in complement component C4 to form C4a and C4b, and Lys(or Arg)
Ala bond in complement component C4 to form C4a and C4b, and Lys(or Arg) Lys bond in complement component C2 to form C2a and C2b: the "classical" pathway C3 convertase
Lys bond in complement component C2 to form C2a and C2b: the "classical" pathway C3 convertase
Other names: C1 esterase; activated complement C1s; complement C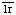
Comments: Activated from proenzyme C1s in plasma by complement subcomponent C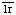 . In peptidase family S1 (trypsin family)
. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 80295-70-1
References
1. Sim, R.B. The human complement system serine proteases C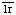 and C
and C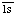 and their proenzymes. Methods Enzymol. 80 (1981) 26-42. [PMID: 6281620]
and their proenzymes. Methods Enzymol. 80 (1981) 26-42. [PMID: 6281620]
2. MacKinnon, C.M., Carter, P.E., Smyth, S.J., Dunbar, B. and Fothergill, J.E. Molecular cloning of cDNA for human complement component C1s. The complete amino acid sequence. Eur. J. Biochem. 169 (1987) 547-553 . [PMID: 3500856]
3. Müller-Eberhard, H.J. Molecular organization and function of the complement system. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
4. Skoog, M.T., Mehdi, S., Wiseman, J.S. and Bey, P. The specificity of two proteinases that cleave adjacent to arginine, C1 esterase and acrosin, for peptide p-nitroanilide substrates. Biochim. Biophys. Acta 996 (1989) 89-94. [PMID: 2500154]
[EC 3.4.21.42 created 1981]
EC 3.4.21.43
Accepted name: classical-complement-pathway C3/C5 convertase
Reaction: Selective cleavage of Arg Ser bond in complement component C3 α-chain to form C3a and C3b, and Arg
Ser bond in complement component C3 α-chain to form C3a and C3b, and Arg bond in complement component C5 α-chain to form C5a and C5b
bond in complement component C5 α-chain to form C5a and C5b
Other names: C3 convertase; C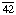 ; C4b,2a; C5 convertase; C
; C4b,2a; C5 convertase; C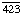 ; C4b,2a,3b; C42; C5 convertase; C423; C4b,2a,3b; complement C.hivin.4.hivin2; complement C3 convertase
; C4b,2a,3b; C42; C5 convertase; C423; C4b,2a,3b; complement C.hivin.4.hivin2; complement C3 convertase
Comments: A complex of complement fragments C4b, C2a and C2b. C2a contains the active site, C2b the site for C4b binding. C2a and C2b are formed by cleavage of proenzyme C2 by complement subcomponent C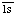 . Cleavage of C5 requires complement fragment C3b which binds C5 and renders it susceptible to cleavage by the C4b,2a complex. Includes former EC 3.4.21.44. Complement component C2a is In peptidase family S1 (trypsin family)
. Cleavage of C5 requires complement fragment C3b which binds C5 and renders it susceptible to cleavage by the C4b,2a complex. Includes former EC 3.4.21.44. Complement component C2a is In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 56626-15-4
References
1. Kerr, M.A. The second component of human complement. Methods Enzymol. 80 (1981) 54-64. [PMID: 7043188]
2. Müller-Eberhard, H.J. Molecular organization and function of the complement system. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
[EC 3.4.21.43 created 1981 (EC 3.4.21.44 created 1981, incorporated 1984)]
[EC 3.4.21.44 Transferred entry: now included with EC 3.4.21.43 - classical-complement-pathway C3/C5 convertase (EC 3.4.21.44 created 1981, deleted 1984)]
EC 3.4.21.45
Accepted name: complement factor I
Reaction: Inactivates complement subcomponents C3b, iC3b and C4b by proteolytic cleavage
Other names: complement component C3b inactivator; C3b inactivator; C3b/C4b inactivator; C3bINA; complement C3b/C4b inactivator; complement C4b inactivator; conglutinogen-activating factor C; complement C3b inactivator; factor I; complement C4bi
Comments: Cleavage of complement subcomponent C3b requires its binding to cofactor factor H or complement receptor CR1; cleavage of iC3b requires complement receptor CR1; cleavage of C4b requires C4b-binding protein. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 80295-66-5
References
1. Nagasawa, S., Ichihara, C. and Stroud, R.M. Cleavage of C4b by C3b inactivator: production of a nicked form from C4b, C4b', as an intermediate cleavage product of C4b by C3b inactivator. J. Immunol. 125 (1980) 578-582. [PMID: 7391570]
2. Crossley, L.G. C3b inactivator and β1H. Methods Enzymol. 80 (1981) 112-124. [PMID: 6210825]
3. Müller-Eberhard, H.J. Molecular organization and function of the complement system. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
[EC 3.4.21.45 created 1981]
EC 3.4.21.46
Accepted name: complement factor D
Reaction: Selective cleavage of Arg Lys bond in complement factor B when in complex with complement subcomponent C3b or with cobra venom factor
Lys bond in complement factor B when in complex with complement subcomponent C3b or with cobra venom factor
Other names: C3 proactivator convertase; properdin factor D esterase; factor D; factor D (complement)
Comments: A component of the alternative pathway of complement activation. This reaction is analogous to the activation of complement component C2 by complement subcomponent C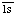 . In peptidase family S1 (trypsin family)
. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 37213-56-2
References
1. Reid, K.B.M., Johnson, D.M.A., Gagnon, J. and Prohaska, R. Preparation of human factor 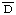 of the alternative pathway of complement. Methods Enzymol. 80 (1981) 134-143. [PMID: 6918766]
of the alternative pathway of complement. Methods Enzymol. 80 (1981) 134-143. [PMID: 6918766]
2. Müller-Eberhard, H.J. Molecular organization and function of the complement system. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
[EC 3.4.21.46 created 1981]
EC 3.4.21.47
Accepted name: alternative-complement-pathway C3/C5 convertase
Reaction: Cleavage of Arg Ser bond in complement component C3 α-chain to yield C3a and C3b, and Arg
Ser bond in complement component C3 α-chain to yield C3a and C3b, and Arg bond in complement component C5 α-chain to yield C5a and C5b
bond in complement component C5 α-chain to yield C5a and C5b
Other names: complement component C3/C5 convertase (alternative); proenzyme factor B; properdin factor B; C3 proactivator; glycine-rich β-glycoprotein; heat-labile factor; C3 convertase; C3b,Bb,CVF,Bb,C5 convertase; (C3b)n,Bb; complement C 3(C 5) convertase (amplification); alternative complement pathway C3(C5) convertase; C5 convertase; CVF,Bb; (CVF)-dependent glycine-rich-beta-glucoprotein; cobra venom factor-dependent C3 convertase
Comments: A bimolecular complex of complement fragment Bb with either C3b or cobra venom factor; Bb contains the active site. Bb is formed by cleavage of proenzyme factor B by factor D. Cleavage of complement component C5 requires additional C3b which binds C5 and renders it susceptible to cleavage by C3b,Bb complex. C3b,Bb is stabilized in plasma by factor P. Complement factor B is In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 80295-67-6
References
1. Kerr, M.A. Human factor B. Methods Enzymol. 80 (1981) 102-112 . [PMID: 7043188]
2. Morley, B.J. and Campbell, R.D. Internal homologies of the Ba fragment from human complement component Factor B, as a class III MHC antigen. EMBO J. 3 (1984) 153-157. [PMID: 6323161]
3. Müller-Eberhard, H.J. The second component of human complement. Annu. Rev. Biochem. 57 (1988) 321-347. [PMID: 3052276]
[EC 3.4.21.47 created 1981]
EC 3.4.21.48
Accepted name: cerevisin
Reaction: Hydrolysis of proteins with broad specificity, and of Bz-Arg-OEt > Ac-Tyr-OEt. Does not hydrolyze peptide amides
Other names: yeast proteinase B; proteinase yscB; baker's yeast proteinase B; brewer's yeast proteinase; peptidase β
Comments: From Saccharomyces cerevisiae (baker's yeast, brewer's yeast). In peptidase family S8 (subtilisin family), but contains a Cys residue near the active site His, and is inhibited by mercurials. Proteinase ycaB is a similar enzyme from the yeast Candida albicans [3]
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
CAS registry number: 37288-81-6
References
1. Félix, F. and Brouillet, N. Purification et propriétés de deux peptidases de levure de brassérie. Biochim. Biophys. Acta 122 (1966) 127-144. [PMID: 4961236]
2. Kominami, E., Hoffschulte, H., Leuschel, L., Maier, K. and Holzer, H. The substrate specificity of proteinase B from baker's yeast. Biochim. Biophys. Acta. 661 (1981) 136-141. [PMID: 7028121]
3. Farley, P.C., Shepherd, M.G. and Sullivan, P.A. The purification and properties of yeast proteinase B from Candida albicans. Biochem. J. 236 (1986) 177-184. [PMID: 3539100]
4. Moehle, C.M., Tizard, R., Lemmon, S.K., Smart, J. and Jones, E.W. Protease B of the lysosome like vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol. Cell. Biol. 7 (1987) 4390-4399. [PMID: 3325823]
[EC 3.4.21.48 created 1972 as EC 3.4.22.9, transferred 1981 to EC 3.4.21.48]
EC 3.4.21.49
Accepted name: hypodermin C
Reaction: Hydrolysis of proteins including native collagen at  Ala bond leaving an N-terminal (75%) and a C-terminal (25%) fragment
Ala bond leaving an N-terminal (75%) and a C-terminal (25%) fragment
Other names: Hypoderma collagenase
Comments: From the larva of a warble fly, Hypoderma lineatum. Little action on small molecule substrates of trypsin, chymotrypsin, elastase or microbial collagenases. In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 122191-36-0
References
1. Lecroisey, A., Boulard, C. and Keil, B. Chemical and enzymatic characterization of the collagenase from the insect Hypoderma lineatum. Eur. J. Biochem. 101 (1979) 385-393. [PMID: 230030]
2. Lecroisey, A. and Keil, B. Specificity of the collagenase from the insect Hypoderma lineatum. Eur. J. Biochem. 152 (1985) 123-130. [PMID: 2995028]
3. Lecroisey, A., Gilles, A.-M., De Wolf, A. and Keil, B. Complete amino acid sequence of the collagenase from the insect Hypoderma lineatum. J. Biol. Chem. 262 (1987) 7546-7551. [PMID: 3034899]
[EC 3.4.21.49 created 1981]
EC 3.4.21.50
Accepted name: lysyl endopeptidase
Reaction: Preferential cleavage: Lys , including -Lys
, including -Lys Pro-
Pro-
Other names: Achromobacter proteinase I (also see Comment); Achromobacter lyticus alkaline proteinase I; protease I; achromopeptidase; lysyl bond specific proteinase
Comments: From Achromobacter lyticus [6]. Enzymes with similar specificity are produced by Lysobacter enzymogenes (Endoproteinase Lys-C; [3]) and Pseudomonas aeruginosa (Ps-1; [4]). In peptidase family S1 (trypsin family)
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MEROPS,
Metacyc,
PDB,
CAS registry number: 123175-82-6
References
1. Masaki, T., Tanabe, M., Nakamura, K. and Soejima, M. Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. I. Purification and some enzymatic properties. Biochim. Biophys. Acta. 660 (1981) 44-50. [PMID: 6791693]
2. Masaki, T., Fujihasi, T., Nakamura, K. and Soejima, M. Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. II. Specificity and inhibition studies of Achromobacter protease I. Biochim. Biophys. Acta 660 (1981) 51-55. [PMID: 6168293]
3. Jekel, P.A., Weijer, W.J. and Beintema, J.J. Use of endoproteinase Lys-C from Lysobacter enzymogenes in protein sequence analysis. Anal. Biochem. 134 (1983) 347-354. [PMID: 6359954]
4. Elliott, B.W. and Cohen, C. Isolation and characterization of a lysine-specific protease from Pseudomonas aeruginosa. J. Biol. Chem. 261 (1986) 11259-11265. [PMID: 3090046]
5. Ohara, T., Makino, K., Shinagawa, H., Nakata, A., Norioka, S. and Sakiyama, F. Cloning, nucleotide sequence, and expression of Achromobacter protease I gene. J. Biol. Chem. 264 (1989) 20625-2063
6. Tsunasawa, S., Masaki, T., Hirose, M., Soejima, M. and Sakiyama, F. The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. J. Biol. Chem. 264 (1989) 3832-3839. [PMID: 2492988]
[EC 3.4.21.50 created 1983]
Continued with EC 3.4.21.51 - EC 3.4.21.105
Return to Peptidases Home Page.
Return to Enzymes Home Page.