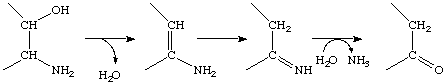
Continued from EC 4.2.99
Others, however, catalyse elimination of another component, e.g. water, which is followed by spontaneous reactions that lead to breakage of the C-N bond, e.g.
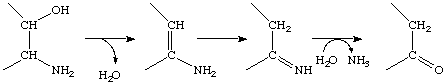
as in EC 4.3.1.17 (L-serine ammonia-lyase), so that the overall reaction is
Sections
EC 4.3.1 Ammonia-Lyases
EC 4.3.2 Lyases Acting on Amides, Amidines, etc.
EC 4.3.3 Amine-Lyases
EC 4.3.99 Other Carbon-Nitrogen Lyases
Accepted name: aspartate ammonia-lyase
Reaction: L-aspartate = fumarate + NH3
Other name(s): aspartase; fumaric aminase; L-aspartase; L-aspartate ammonia-lyase
Systematic name: L-aspartate ammonia-lyase (fumarate-forming)
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9027-30-9
References:
1. Ellfolk, N. Studies on aspartase. 1. Quantitative separation of aspartase from bacterial cells, and its partial purification. Acta Chem. Scand. 7 (1953) 824-830.
Accepted name: methylaspartate ammonia-lyase
Reaction: L-threo-3-methylaspartate = mesaconate + NH3
Other name(s): β-methylaspartase; 3-methylaspartase; L-threo-3-methylaspartate ammonia-lyase
Systematic name: L-threo-3-methylaspartate ammonia-lyase (mesaconate-forming)
Comments: A cobalamin protein.
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9033-26-5
References:
1. Barker, H.A., Smyth, R.O., Wawszkiewicz, E.J., Lee, M.N. and Wilson, R.M. Enzymic preparation and characterization of an α-L-β-methylaspartic acid. Arch. Biochem. Biophys. 78 (1958) 468-476.
2. Bright, H.J. and Ingraham, L.L. The preparation of crystalline β-methylaspartase. Biochim. Biophys. Acta 44 (1960) 586-588. [PMID: 13618029]
Accepted name: histidine ammonia-lyase
Reaction: L-histidine = urocanate + NH3
For diagram of reaction click here
Glossary: urocanate = (E)-3-(imidazol-4-yl)propenoate
Other name(s): histidase; histidinase; histidine α-deaminase; L-histidine ammonia-lyase
Systematic name: L-histidine ammonia-lyase (urocanate-forming)
Comments: This enzyme is a member of the aromatic amino acid lyase family, other members of which are EC 4.3.1.23 (tyrosine ammonia-lyase), EC 4.3.1.24 (phenylalanine ammonia-lyase) and EC 4.3.1.25 (phenylalanine/tyrosine ammonia-lyase). The enzyme contains the cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO), which is common to this family [4]. This unique cofactor is formed autocatalytically by cyclization and dehydration of the three amino-acid residues alanine, serine and glycine [5]. This enzyme catalyses the first step in the degradation of histidine and the product, urocanic acid, is further metabolized to glutamate [2,3].
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9013-75-6
References:
1. Mehler, A.H. and Tabor, H. Deamination of histidine to form urocanic acid in liver. J. Biol. Chem. 201 (1953) 775-784. [PMID: 13061415]
2. Watts, K.T., Mijts, B.N., Lee, P.C., Manning, A.J. and Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13 (2006) 1317-1326. [PMID: 17185227]
3. Poppe, L. and Rétey, J. Friedel-Crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine. Angew. Chem. Int. Ed. Engl. 44 (2005) 3668-3688. [PMID: 15906398]
4. Louie, G.V., Bowman, M.E., Moffitt, M.C., Baiga, T.J., Moore, B.S. and Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 13 (2006) 1327-1338. [PMID: 17185228]
5. Schwede, T.F., Rétey, J. and Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38 (1999) 5355-5361. [PMID: 10220322]
Accepted name: formimidoyltetrahydrofolate cyclodeaminase
Reaction: 5-formimidoyltetrahydrofolate = 5,10-methenyltetrahydrofolate + NH3
For diagram of reaction click here (another example).
Other name(s): formiminotetrahydrofolate cyclodeaminase; 5-formimidoyltetrahydrofolate ammonia-lyase (cyclizing)
Systematic name: 5-formimidoyltetrahydrofolate ammonia-lyase (cyclizing; 5,10-methenyltetrahydrofolate-forming)
Comments: In eukaroytes, occurs as a bifunctional enzyme that also has glutamate formimidoyltransferase (EC 2.1.2.5) activity.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9032-05-7
References:
1. Rabinowitz, J.C. and Pricer, W.E. Formimino-tetrahydrofolic acid and methenyltetrahydrofolic acid as intermediates in the formation of N10-formyltetrahydrofolic acid. J. Am. Chem. Soc. 78 (1956) 5702-5704.
[EC 4.3.1.5 Transferred entry: phenylalanine ammonia-lyase. Now divided into EC 4.3.1.23 (tyrosine ammonia-lyase), EC 4.3.1.24 (phenylalanine ammonia-lyase) and EC 4.3.1.25 (phenylalanine/tyrosine ammonia-lyase). (EC 4.3.1.5 created 1965, deleted 2008)]
Accepted name: β-alanyl-CoA ammonia-lyase
Reaction: β-alanyl-CoA = acryloyl-CoA + NH3
Other name(s): β-alanyl coenzyme A ammonia-lyase; β-alanyl-CoA ammonia-lyase
Systematic name: β-alanyl-CoA ammonia-lyase (acryloyl-CoA-forming)
Comments: The reaction has only been demonstrated in the direction of addition of ammonia.
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, CAS registry number: 9024-29-7
References:
1. Stadtman, E.R. The enzymic synthesis of β-alanyl coenzyme A. J. Am. Chem. Soc. 77 (1955) 5765-5766.
Accepted name: ethanolamine ammonia-lyase
Reaction: ethanolamine = acetaldehyde + NH3
Other name(s): ethanolamine deaminase; ethanolamine ammonia-lyase
Systematic name: ethanolamine ammonia-lyase (acetaldehyde-forming)
Comments: A cobalamin protein.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9054-69-7
References:
1. Bradbeer, C. The clostridial fermentations of choline and ethanolamine. 1. Preparation and properties of cell-free extracts. J. Biol. Chem. 240 (1965) 4669-4474. [PMID: 5846987]
2. Bradbeer, C. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J. Biol. Chem. 240 (1965) 4675-4681. [PMID: 5846988]
3. Kaplan, B.H. and Stadtman, E.R. Ethanolamine deaminase, a cobamide coenzyme-dependent enzyme. I. Purification, assay, and properties of the enzyme. J. Biol. Chem. 243 (1968) 1787-1793. [PMID: 4297225]
[EC 4.3.1.8 Transferred entry: now EC 2.5.1.61 hydroxymethylbilane synthase. (EC 4.3.1.8 created 1972, deleted 2003)]
Accepted name: glucosaminate ammonia-lyase
Reaction: 2-amino-2-deoxy-D-gluconate = 2-dehydro-3-deoxy-D-gluconate + NH3 (overall reaction)
(1a) 2-amino-2-deoxy-D-gluconate = (2Z,4S,5R)-2-amino-4,5,6-trihydroxyhex-2-enoate + H2O
(1b) (2Z,4S,5R)-2-amino-4,5,6-trihydroxyhex-2-enoate = (4S,5R)-4,5,6-trihydroxy-2-iminohexanoate (spontaneous)
(1c) (4S,5R)-4,5,6-trihydroxy-2-iminohexanoate + H2O = 2-dehydro-3-deoxy-D-gluconate + NH3 (spontaneous)
Glossary: 2-amino-2-deoxy-D-gluconate = glucosaminate
Other name(s): glucosaminic dehydrase; D-glucosaminate dehydratase; D-glucosaminic acid dehydrase; aminodeoxygluconate dehydratase; 2-amino-2-deoxy-D-gluconate hydro-lyase (deaminating); aminodeoxygluconate ammonia-lyase; 2-amino-2-deoxy-D-gluconate ammonia-lyase; D-glucosaminate ammonia-lyase
Systematic name: D-glucosaminate ammonia-lyase (isomerizing; 2-dehydro-3-deoxy-D-gluconate-forming)
Comments: Contains pyridoxal phosphate. The enzyme releases an unstable enamine product that tautomerizes to an imine form, which undergoes spontaneous hydrolytic deamination to form the final product.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 37290-91-8
References:
1. Imanaga, Y. Metabolism of D-glucosamine. III. Enzymic degradation of D-glucosaminic acid. J. Biochem. (Tokyo) 45 (1958) 647-650.
2. Merrick, J.M. and Roseman, S. D-Glucosaminic acid dehydrase. Methods Enzymol. 9 (1966) 657-660.
3. Iwamoto, R., Imanaga, Y. and Soda, K. D-Glucosaminate dehydratase from Agrobacterium radiobacter. Physicochemical and enzymological properties. J. Biochem. (Tokyo) 91 (1982) 283-289. [PMID: 7068563]
4. Iwamoto, R., Taniki, H., Koishi, J. and Nakura, S. D-Glucosaminate aldolase activity of D-glucosaminate dehydratase from Pseudomonas fluorescens and its requirement for Mn2+ ion. Biosci. Biotechnol. Biochem. 59 (1995) 408-411. [PMID: 7766176]
Accepted name: serine-sulfate ammonia-lyase
Reaction: L-serine O-sulfate + H2O = pyruvate + NH3 + sulfate
Other name(s): (L-SOS)lyase
Systematic name: L-serine-O-sulfate ammonia-lyase (pyruvate-forming)
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 9054-70-0
References:
1. Thomas, J.H. and Tudball, N. Studies on the enzymic degradation of L-serine O-sulphate by a rat liver preparation. Biochem. J. 105 (1967) 467-472. [PMID: 5583990]
[EC 4.3.1.11 Deleted entry: dihydroxyphenylalanine ammonia-lyase. The entry had been drafted on the basis of a single abstract that did not provide experimental evidence of the enzyme-catalysed reaction. (EC 4.3.1.11 created 1972, deleted 2007)]
Accepted name: ornithine cyclodeaminase
Reaction: L-ornithine = L-proline + NH3
For diagram click here.
Other name(s): ornithine cyclase; ornithine cyclase (deaminating); L-ornithine ammonia-lyase (cyclizing)
Systematic name: L-ornithine ammonia-lyase (cyclizing; L-proline-forming)
Comments: Requires NAD+. The enzyme is a member of the μ-crystallin protein family [4]. The reaction is stimulated by the presence of ADP or ATP and is inhibited by O2 [2].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9054-76-6
References:
1. Costilow, R.N. and Laycock, L. Ornithine cyclase (deaminating). Purification of a protein that converts ornithine to proline and definition of the optimal assay conditions. J. Biol. Chem. 246 (1971) 6655-6660. [PMID: 4399881]
2. Muth, W.L. and Costilow, R.N. Ornithine cyclase (deaminating). II. Properties of the homogeneous enzyme. J. Biol. Chem. 249 (1974) 7457-7462. [PMID: 4373469]
3. Espineda, C.E., Linford, A.S., Devine, D. and Brusslan, J.A. The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 (1999) 10507-10511. [PMID: 10468639]
4. Goodman, J.L., Wang, S., Alam, S., Ruzicka, F.J., Frey, P.A. and Wedekind, J.E. Ornithine cyclodeaminase: structure, mechanism of action, and implications for the μ-crystallin family. Biochemistry 43 (2004) 13883-13891. [PMID: 15518536]
5. Alam, S., Wang, S.C., Ruzicka, F.J., Frey, P.A. and Wedekind, J.E. Crystallization and X-ray diffraction analysis of ornithine cyclodeaminase from Pseudomonas putida. Acta Crystallogr. D Biol. Crystallogr. 60 (2004) 941-944. [PMID: 15103146]
Accepted name: carbamoyl-serine ammonia-lyase
Reaction: O-carbamoyl-L-serine + H2O = pyruvate + 2 NH3 + CO2 (overall reaction)
(1a) O-carbamoyl-L-serine = CO2 + NH3 + 2-aminoprop-2-enoate
(1b) 2-aminoprop-2-enoate = 2-iminopropanoate (spontaneous)
(1c) 2-iminopropanoate + H2O = pyruvate + NH3 (spontaneous)
Other name(s): O-carbamoyl-L-serine deaminase; carbamoylserine deaminase; O-carbamoyl-L-serine ammonia-lyase (pyruvate-forming)
Systematic name: O-carbamoyl-L-serine ammonia-lyase (decarboxylating; pyruvate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme cleaves a carbon-oxygen bond, releasing CO2, ammonia, and an unstable enamine product that tautomerizes to an imine form, which undergoes a hydrolytic deamination to form pyruvate and a second ammonia molecule. The latter reaction, which can occur spontaneously, can also be catalysed by EC 3.5.99.10, 2-iminobutanoate/2-iminopropanoate deaminase.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 52227-64-2
References:
1. Cooper, A.J.L. and Meister, A. Enzymatic conversion of O-carbamyl-L-serine to pyruvate and ammonia. Biochem. Biophys. Res. Commun. 55 (1973) 780-787. [PMID: 4399881]
Accepted name: 3-aminobutyryl-CoA ammonia-lyase
Reaction: L-3-aminobutyryl-CoA = crotonoyl-CoA + NH3
For diagram of reaction click here.
Other name(s): L-3-aminobutyryl-CoA deaminase; L-3-aminobutyryl-CoA ammonia-lyase
Systematic name: L-3-aminobutyryl-CoA ammonia-lyase (crotonoyl-CoA-forming)
Comments: Hydroxylamine can replace ammonia as a substrate. Crotonoyl-pantetheine can replace crotonoyl-CoA but it is a poorer substrate.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 55467-41-9
References:
1. Jeng, I.-M., Barker, H.A. Purification and properties of L-3-aminobutyryl coenzyme A deaminase from a lysine-fermenting Clostridium. J. Biol. Chem. 249 (1974) 6578-6584. [PMID: 4420467]
2. Barker, H.A., Kahn, J.M., Chew, S. Enzymes involved in 3,5-diaminohexanoate degradation by Brevibacterium sp. J. Bacteriol. 143 (1980) 1165-1170. [PMID: 7410315]
Accepted name: diaminopropionate ammonia-lyase
Reaction: 2,3-diaminopropanoate + H2O = pyruvate + 2 NH3
Other name(s): diaminopropionatase; α,β-diaminopropionate ammonia-lyase; 2,3-diaminopropionate ammonia-lyase; 2,3-diaminopropanoate ammonia-lyase
Systematic name: 2,3-diaminopropanoate ammonia-lyase (adding water; pyruvate-forming)
Comments: A pyridoxal phosphate enzyme. Active towards both D- and L-diaminopropanoate. D- and L-serine are poor substrates.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 51901-19-0
References:
1. Nagasawa, T., Tanizawa, K., Satoda, T., Yamada, H. Diaminopropionate ammonia-lyase from Salmonella typhimurium. Purification and characterization of the crystalline enzyme, and sequence determination of the pyridoxal 5'-phosphate binding peptide. J. Biol. Chem. 263 (1988) 958-964. [PMID: 3275662]
Accepted name: threo-3-hydroxy-L-aspartate ammonia-lyase
Reaction: threo-3-hydroxy-L-aspartate = oxaloacetate + NH3
Other name(s): L-threo-3-hydroxyaspartate dehydratase; threo-3-hydroxyaspartate ammonia-lyase
Systematic name: threo-3-hydroxy-L-aspartate ammonia-lyase (oxaloacetate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme, purified from the bacterium Pseudomonas sp. T62, is highly specific, and does not accept any other stereoisomer of 3-hydroxyaspartate. Different from EC 4.3.1.20, erythro-3-hydroxy-L-aspartate ammonia-lyase and EC 4.3.1.27, threo-3-hydroxy-D-aspartate ammonia-lyase. Requires a divalent cation such as Mn2+, Mg2+, or Ca2+.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 248270-70-4
References:
1. Wada, M., Matsumoto, T., Nakamori, S., Sakamoto, M., Kataoka, M., Liu, J.-Q., Itoh, N., Yamada, H. and Shimizu, S. Purification and characterization of a novel enzyme, L-threo-3-hydroxyaspartate dehydratase, from Pseudomonas sp. T62. FEMS Microbiol. Lett. 179 (1999) 147-151. [PMID: 10481099]
Accepted name: L-serine ammonia-lyase
Reaction: L-serine = pyruvate + NH3 (overall reaction)
(1a) L-serine = 2-aminoprop-2-enoate + H2O
(1b) 2-aminoprop-2-enoate = 2-iminopropanoate (spontaneous)
(1c) 2-iminopropanoate + H2O = pyruvate + NH3 (spontaneous)
Other name(s): serine deaminase; L-hydroxyaminoacid dehydratase; L-serine deaminase; L-serine dehydratase; L-serine hydro-lyase (deaminating)
Systematic name: L-serine ammonia-lyase (pyruvate-forming)
Comments: Most enzymes that catalyse this reaction are pyridoxal-phosphate-dependent, although some enzymes contain an iron-sulfur cluster instead [6]. The reaction catalysed by both types of enzymes involves the initial elimination of water to form an enamine intermediate (hence the enzyme’s original classification as EC 4.2.1.13, L-serine dehydratase), followed by tautomerization to an imine form and hydrolysis of the C-N bond. The latter reaction, which can occur spontaneously, is also be catalysed by EC 3.5.99.10, 2-iminobutanoate/2-iminopropanoate deaminase. This reaction is also carried out by EC 4.3.1.19, threonine ammonia-lyase, from a number of sources.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number: 9014-27-1
References:
1. Ramos, F. and Wiame, J.-M. Occurrence of a catabolic L-serine (L-threonine) deaminase in Saccharomyces cerevisiae. Eur. J. Biochem. 123 (1982) 571-576. [PMID: 7042346]
2. Simon, D., Hoshino, J. and Kröger, H. L-Serine dehydratase from rat liver. Purification and some properties. Biochim. Biophys. Acta 321 (1973) 361-368. [PMID: 4750769]
3. Suda, M. and Nakagawa, H. L-Serine dehydratase (rat liver). Methods Enzymol. 17B (1971) 346-351.
4. Sagers, R.D. and Carter, J. E. L-Serine dehydratase (Clostridium acidiurica). Methods Enzymol. 17B (1971) 351-356.
5. Robinson, W.G. and Labow, R. L-Serine dehydratase (Escherichia coli). Methods Enzymol. 17B (1971) 356-360.
6. Grabowski, R., Hofmeister, A.E. and Buckel, W. Bacterial L-serine dehydratases: a new family of enzymes containing iron-sulfur clusters. Trends Biochem. Sci. 18 (1993) 297-300. [PMID: 8236444]
7. Yamada, T., Komoto, J., Takata, Y., Ogawa, H., Pitot, H.C. and Takusagawa, F. Crystal structure of serine dehydratase from rat liver. Biochemistry 42 (2003) 12854-12865. [PMID: 14596599]
Accepted name: D-serine ammonia-lyase
Reaction: D-serine = pyruvate + NH3 (overall reaction)
(1a) D-serine = 2-aminoprop-2-enoate + H2O
(1b) 2-aminoprop-2-enoate = 2-iminopropanoate (spontaneous)
(1c) 2-iminopropanoate + H2O = pyruvate + NH3 (spontaneous)
Other name(s): D-hydroxyaminoacid dehydratase; D-serine dehydrase; D-hydroxy amino acid dehydratase; D-serine hydrolase; D-serine dehydratase (deaminating); D-serine deaminase; D-serine hydro-lyase (deaminating)
Systematic name: D-serine ammonia-lyase (pyruvate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme cleaves a carbon-oxygen bond, releasing a water molecule (hence the enzyme’s original classification as EC 4.2.1.14, D-serine dehydratase) and an unstable enamine product that tautomerizes to an imine form, which undergoes a hydrolytic deamination to form pyruvate and ammonia. The latter reaction, which can occur spontaneously, can also be catalysed by EC 3.5.99.10, 2-iminobutanoate/2-iminopropanoate deaminase. Also acts, slowly, on D-threonine.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9015-88-7
References:
1. Dupourque, D., Newton, W.A. and Snell, E.E. Purification and properties of D-serine dehydrase from Escherichia coli. J. Biol. Chem. 241 (1966) 1233-1238. [PMID: 5327100]
2. Metzler, D.E. and Snell, E.E. Deamination of serine. II. D-Serine dehydrase, a vitamin B6 enzyme from Escherichia coli. J. Biol. Chem. 198 (1952) 363-373. [PMID: 12999751]
Accepted name: threonine ammonia-lyase
Reaction: L-threonine = 2-oxobutanoate + NH3 (overall reaction)
(1a) L-threonine = 2-aminobut-2-enoate + H2O
(1b) 2-aminobut-2-enoate = 2-iminobutanoate (spontaneous)
(1c) 2-iminobutanoate + H2O = 2-oxobutanoate + NH3 (spontaneous)
For diagram of reaction click here.
Other name(s): threonine deaminase; L-serine dehydratase; serine deaminase; L-threonine dehydratase; threonine dehydrase; L-threonine deaminase; threonine dehydratase; L-threonine hydro-lyase (deaminating); L-threonine ammonia-lyase
Systematic name: L-threonine ammonia-lyase (2-oxobutanoate-forming)
Comments: Most enzymes that catalyse this reaction are pyridoxal-phosphate-dependent, although some enzymes contain an iron-sulfur cluster instead. The reaction catalysed by both types of enzymes involves the initial elimination of water to form an enamine intermediate (hence the enzyme's original classification as EC 4.2.1.16, threonine dehydratase), followed by tautomerization to an imine form and hydrolysis of the C-N bond [3,5]. The latter reaction, which can occur spontaneously, is also be catalysed by EC 3.5.99.10, 2-iminobutanoate/2-iminopropanoate deaminase [5]. The enzymes from a number of sources also act on L-serine, cf. EC 4.3.1.17, L-serine ammonia-lyase.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, MetaCyc, PDB, CAS registry number: 774231-81-1
References:
1. Cohn, M.S. and Phillips, A.T. Purification and characterization of a B6-independent threonine dehydratase from Pseudomonas putida. Biochemistry 13 (1974) 1208-1214. [PMID: 4814721]
2. Nishimura, J.S. and Greenberg, D.M. Purification and properties of L-threonine dehydrase of sheep liver. J. Biol. Chem. 236 (1961) 2684-2691. [PMID: 14479973]
3. Phillips, A.T. and Wood, W.A. The mechanism of action of 5'-adenylic acid-activated threonine dehydrase. J. Biol. Chem. 240 (1965) 4703-4709. [PMID: 5321308]
4. Shizuta, Y., Nakazawa, A., Tokushige, M. and Hayaishi, O. Studies on the interaction between regulatory enzymes and effectors. 3. Crystallization and characterization of adenosine 5'-monophosphate-dependent threonine deaminase from Escherichia coli. J. Biol. Chem. 244 (1969) 1883-1889. [PMID: 4889010]
5. Lambrecht, J.A., Flynn, J.M. and Downs, D.M. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5'-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 287 (2012) 3454-3461. [PMID: 22094463]
Accepted name: erythro-3-hydroxy-L-aspartate ammonia-lyase
Reaction: erythro-3-hydroxy-L-aspartate = oxaloacetate + NH3
Other name(s): erythro-β-hydroxyaspartate dehydratase; erythro-3-hydroxyaspartate dehydratase; erythro-3-hydroxy-Ls-aspartate hydro-lyase (deaminating); erythro-3-hydroxy-Ls-aspartate ammonia-lyase
Systematic name: erythro-3-hydroxy-L-aspartate ammonia-lyase (oxaloacetate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme, which was characterized from the bacterium Paracoccus denitrificans NCIMB 8944, is highly specific for the L-isomer of erythro-3-hydroxyaspartate. Different from EC 4.3.1.16, threo-3-hydroxy-L-aspartate ammonia-lyase and EC 4.3.1.27, threo-3-hydroxy-D-aspartate ammonia-lyase. Requires a divalent cation such as Mn2+, Mg2+, and Ca2+.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 37290-74-7
References:
1. Gibbs, R.G. and Morris, J.G. Purification and properties of erythro-β-hydroxyaspartate dehydratase from Micrococcus denitrificans. Biochem. J. 97 (1965) 547-554. [PMID: 16749162]
[EC 4.3.1.21 Deleted entry: aminodeoxygluconate ammonia-lyase. Enzyme is identical to EC 4.3.1.9, glucosaminate ammonia-lyase (EC 4.3.1.21 created 1965 as EC 4.2.1.26, transferred 2002 to EC 4.3.1.21, deleted 2004)]
Accepted name: 3,4-dihydroxyphenylalanine reductive deaminase
Reaction: L-dopa + NADH = 3,4-dihydroxyphenylpropanoate + NAD+ + NH3
Glossary: L-dopa = 3,4-dihydroxy-L-phenylalanine
Other name(s): reductive deaminase; DOPA-reductive deaminase; DOPARDA
Systematic name: 3,4-dihydroxy-L-phenylalanine ammonia-lyase (3,4-dihydroxyphenylpropanoate-forming)
Comments: Forms part of the L-phenylalanine-catabolism pathway in the anoxygenic phototrophic bacterium Rhodobacter sphaeroides OU5. NADPH is oxidized more slowly than NADH.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Ranjith, N.K., Sasikala, Ch. and Ramana, Ch.V. Catabolism of L-phenylalanine and L-tyrosine by Rhodobacter sphaeroides OU5 occurs through 3,4-dihydroxyphenylalanine. Res. Microbiol. 158 (2007) 506-511. [PMID: 17616348]
Accepted name: tyrosine ammonia-lyase
Reaction: L-tyrosine = trans-p-hydroxycinnamate + NH3
Other name(s): TAL; tyrase; L-tyrosine ammonia-lyase
Systematic name: L-tyrosine ammonia-lyase (trans-p-hydroxycinnamate-forming)
Comments: This enzyme is a member of the aromatic amino acid lyase family, other members of which are EC 4.3.1.3 (histidine ammonia-lyase), EC 4.3.1.24 (phenylalanine ammonia-lyase) and EC 4.3.1.25 (phenylalanine/tyrosine ammonia-lyase). The enzyme contains the cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO), which is common to this family [1]. This unique cofactor is formed autocatalytically by cyclization and dehydration of the three amino-acid residues alanine, serine and glycine [3]. The enzyme is far more active with tyrosine than with phenylalanine as substrate, but the substrate specificity can be switched by mutation of a single amino acid (H89F) in the enzyme from the bacterium Rhodobacter sphaeroides [1,2].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 1030840-68-6
References:
1. Louie, G.V., Bowman, M.E., Moffitt, M.C., Baiga, T.J., Moore, B.S. and Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 13 (2006) 1327-1338. [PMID: 17185228]
2. Watts, K.T., Mijts, B.N., Lee, P.C., Manning, A.J. and Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13 (2006) 1317-1326. [PMID: 17185227]
3. Schwede, T.F., Rétey, J. and Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38 (1999) 5355-5361. [PMID: 10220322]
Accepted name: phenylalanine ammonia-lyase
Reaction: L-phenylalanine = trans-cinnamate + NH3
For diagram of reaction click here
Other name(s): phenylalanine deaminase; phenylalanine ammonium-lyase; PAL; L-phenylalanine ammonia-lyase; Phe ammonia-lyase
Systematic name: L-phenylalanine ammonia-lyase (trans-cinnamate-forming)
Comments: This enzyme is a member of the aromatic amino acid lyase family, other members of which are EC 4.3.1.3 (histidine ammonia-lyase) and EC 4.3.1.23 (tyrosine ammonia-lyase) and EC 4.3.1.25 (phenylalanine/tyrosine ammonia-lyase). The enzyme contains the cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO), which is common to this family [3]. This unique cofactor is formed autocatalytically by cyclization and dehydration of the three amino-acid residues alanine, serine and glycine [9]. The enzyme from some species is highly specific for phenylalanine [7,8].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9024-28-6
References:
1. Koukol, J. and Conn, E.E. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 236 (1961) 2692-2698. [PMID: 14458851]
2. Young, M.R. and Neish, A.C. Properties of the ammonia-lyases deaminating phenylalanine and related compounds in Triticum sestivum and Pteridium aquilinum. Phytochemistry 5 (1966) 1121-1132.
3. Louie, G.V., Bowman, M.E., Moffitt, M.C., Baiga, T.J., Moore, B.S. and Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 13 (2006) 1327-1338. [PMID: 17185228]
4. Calabrese, J.C., Jordan, D.B., Boodhoo, A., Sariaslani, S. and Vannelli, T. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry 43 (2004) 11403-11416. [PMID: 15350127]
5. Ritter, H. and Schulz, G.E. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 16 (2004) 3426-3436. [PMID: 15548745]
6. Watts, K.T., Mijts, B.N., Lee, P.C., Manning, A.J. and Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13 (2006) 1317-1326. [PMID: 17185227]
7. Appert, C., Logemann, E., Hahlbrock, K., Schmid, J. and Amrhein, N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.). Eur. J. Biochem. 225 (1994) 491-499. [PMID: 7925471]
8. Cochrane, F.C., Davin, L.B. and Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65 (2004) 1557-1564. [PMID: 15276452]
9. Schwede, T.F., Rétey, J. and Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38 (1999) 5355-5361. [PMID: 10220322]
Accepted name: phenylalanine/tyrosine ammonia-lyase
Reaction: (1) L-phenylalanine = trans-cinnamate + NH3
(2) L-tyrosine = trans-p-hydroxycinnamate + NH3
Other name(s): PTAL; bifunctional PAL
Systematic name: L-phenylalanine(or L-tyrosine):trans-cinnamate(or trans-p-hydroxycinnamate) ammonia-lyase
Comments: This enzyme is a member of the aromatic amino acid lyase family, other members of which are EC 4.3.1.3 (histidine ammonia-lyase), EC 4.3.1.23 (tyrosine ammonia-lyase) and EC 4.3.1.24 (phenylalanine ammonia-lyase). The enzyme from some monocots, including maize, and from the yeast Rhodosporidium toruloides, deaminate L-phenylalanine and L-tyrosine with similar catalytic efficiency [3]. The enzyme contains the cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO), which is common to this family [3]. This unique cofactor is formed autocatalytically by cyclization and dehydration of the three amino-acid residues alanine, serine and glycine [4].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Rösler, J., Krekel, F., Amrhein, N. and Schmid, J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113 (1997) 175-179. [PMID: 9008393]
2. Watts, K.T., Mijts, B.N., Lee, P.C., Manning, A.J. and Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13 (2006) 1317-1326. [PMID: 17185227]
3. Louie, G.V., Bowman, M.E., Moffitt, M.C., Baiga, T.J., Moore, B.S. and Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 13 (2006) 1327-1338. [PMID: 17185228]
4. Schwede, T.F., Rétey, J. and Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38 (1999) 5355-5361. [PMID: 10220322]
[EC 4.3.1.26 Transferred entry: chromopyrrolate synthase. Now EC 1.21.3.9, dichlorochromopyrrolate synthase (EC 4.3.1.26 created 2010, deleted 2013)]
Accepted name: threo-3-hydroxy-D-aspartate ammonia-lyase
Reaction: threo-3-hydroxy-D-aspartate = oxaloacetate + NH3
Other name(s): D-threo-3-hydroxyaspartate dehydratase
Systematic name: threo-3-hydroxy-D-aspartate ammonia-lyase (oxaloacetate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme, purified from the bacterium Delftia sp. HT23, also has activity against L-threo-3-hydroxyaspartate, L-erythro-3-hydroxyaspartate, and D-serine. Different from EC 4.3.1.20, erythro-3-hydroxy-L-aspartate ammonia-lyase and EC 4.3.1.16, threo-3-hydroxy-L-aspartate ammonia-lyase. Requires a divalent cation such as Mn2+, Co2+ or Ni2+.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Maeda, T., Takeda, Y., Murakami, T., Yokota, A. and Wada, M. Purification, characterization and amino acid sequence of a novel enzyme, D-threo-3-hydroxyaspartate dehydratase, from Delftia sp. HT23. J. Biochem. 148 (2010) 705-712. [PMID: 20843822]
Accepted name: L-lysine cyclodeaminase
Reaction: L-lysine = L-pipecolate + NH3
Other name(s): rapL (gene name); fkbL (gene name); tubZ (gene name); visC (gene name)
Systematic name: L-lysine ammonia-lyase (cyclizing; ammonia-forming)
Comments: Requires bound NAD+. The enzyme produces the non-proteinogenic amino acid L-pipecolate, which is incorporated into multiple secondary metabolite products, including rapamycin, tobulysin, virginiamycin and pristinamycin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Khaw, L.E., Bohm, G.A., Metcalfe, S., Staunton, J. and Leadlay, P.F. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180 (1998) 809-814. [PMID: 9473033]
2. Gatto, G.J., Jr., Boyne, M.T., 2nd, Kelleher, N.L. and Walsh, C.T. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128 (2006) 3838-3847. [PMID: 16536560]
3. Tsotsou, G.E. and Barbirato, F. Biochemical characterisation of recombinant Streptomyces pristinaespiralis L-lysine cyclodeaminase. Biochimie 89 (2007) 591-604. [PMID: 17291665]
Accepted name: D-glucosaminate-6-phosphate ammonia-lyase
Reaction: 2-amino-2-deoxy-D-gluconate 6-phosphate = 2-dehydro-3-deoxy-6-phospho-D-gluconate + NH3
Other name(s): DgaE; 6-phospho-D-glucosaminate ammonia-lyase (2-dehydro-3-deoxy-6-phospho-D-gluconate-forming)
Systematic name: 2-amino-2-deoxy-D-gluconate 6-phosphate ammonia-lyase (2-dehydro-3-deoxy-6-phospho-D-gluconate-forming)
Comments: The enzyme, from the bacterium Salmonella typhimurium, is involved in the degradation pathway of 2-amino-2-deoxy-D-gluconate.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Miller, K.A., Phillips, R.S., Mrazek, J. and Hoover, T.R. Salmonella utilizes D-glucosaminate via a mannose family phosphotransferase system permease and associated enzymes. J. Bacteriol. 195 (2013) 4057-4066. [PMID: 23836865]
Accepted name: dTDP-4-amino-4,6-dideoxy-D-glucose ammonia-lyase
Reaction: dTDP-4-amino-4,6-dideoxy-α-D-glucopyranose + S-adenosyl-L-methionine + reduced acceptor = dTDP-3-dehydro-4,6-dideoxy-α-D-glucopyranose + NH3 + L-methionine + 5'-deoxyadenosine + acceptor
For diagram of reaction click here.
Other name(s): desII (gene name); eryCV (gene name); MegCV
Systematic name: dTDP-4-amino-4,6-dideoxy-α-D-glucopyranose ammonia lyase (dTDP-3-dehydro-4,6-dideoxy-α-D-glucopyranose-forming)
Comments: The enzyme, which is a member of the 'AdoMet radical' (radical SAM) family, is involved in biosynthesis of TDP-α-D-desosamine. The reaction starts by the transfer of an electron from the reduced form of the enzyme's [4Fe-4S] cluster to S-adenosyl-L-methionine, spliting it into methionine and the radical 5-deoxyadenosin-5'-yl, which attacks the sugar substrate.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Szu, P.H., Ruszczycky, M.W., Choi, S.H., Yan, F. and Liu, H.W. Characterization and mechanistic studies of DesII: a radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine. J. Am. Chem. Soc. 131 (2009) 14030-14042. [PMID: 19746907]
2. Ruszczycky, M.W., Choi, S.H. and Liu, H.W. Stoichiometry of the redox neutral deamination and oxidative dehydrogenation reactions catalyzed by the radical SAM enzyme DesII. J. Am. Chem. Soc. 132 (2010) 2359-2369. [PMID: 20121093]
3. Ruszczycky, M.W., Choi, S.H., Mansoorabadi, S.O. and Liu, H.W. Mechanistic studies of the radical S-adenosyl-L-methionine enzyme DesII: EPR characterization of a radical intermediate generated during its catalyzed dehydrogenation of TDP-D-quinovose. J. Am. Chem. Soc. 133 (2011) 7292-7295. [PMID: 21513273]
Accepted name: L-tryptophan ammonia lyase
Reaction: L-tryptophan = 3-indoleacrylate + NH3
Glossary: 3-indoleacrylate = (2E)-3-(1H-indol-3-yl)prop-2-enoate
Other name(s): WAL
Systematic name: L-tryptophan ammonia-lyase (3-indoleacrylate-forming)
Comments: The enzyme, characterized from the bacterium Rubrivivax benzoatilyticus JA2, requires no cofactors. It acts on L-phenylalanine and L-glutamate with about 60% of the activity with L-tryptophan, and on L-tyrosine, glycine, and L-alanine with about 30% of the activity.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Kumavath, R.N., Ramana ChV., Sasikala Ch, Barh, D., Kumar, A.P. and Azevedo, V. Isolation and characterization of L-tryptophan ammonia lyase from Rubrivivax benzoatilyticus strain JA2. Curr Protein Pept Sci 16 (2015) 775-781. [PMID: 25961404]
Accepted name: 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase
Reaction: 5-amino-5-(4-hydroxybenzyl)-6-(D-ribitylimino)-5,6-dihydrouracil + S-adenosyl-L-methionine = 7,8-didemethyl-8-hydroxy-5-deazariboflavin + NH3 + L-methionine + 5'-deoxyadenosine
For diagram of reaction click here.
Glossary: 7,8-didemethyl-8-hydroxy-5-deazariboflavin = Fo
Other name(s): Fo synthase; fbiC (gene name) (ambiguous); cofG (gene name)
Systematic name: 5-amino-5-(4-hydroxybenzyl)-6-(D-ribitylimino)-5,6-dihydrouracil ammonia-lyase (7,8-didemethyl-8-hydroxy-5-deazariboflavin-forming)
Comments: The enzyme produces the Fo precursor of the redox cofactor coenzyme F420, which is found in methanogens and in various actinobacteria. Fo is also produced by some cyanobacteria and eukaryotes. The enzyme, which forms a complex with EC 2.5.1.147, 5-amino-6-(D-ribitylamino)uracil—L-tyrosine 4-hydroxyphenyl transferase, is a radical SAM enzyme that uses the 5'-deoxyadenosyl radical to catalyse the condensation reaction.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Decamps, L., Philmus, B., Benjdia, A., White, R., Begley, T.P. and Berteau, O. Biosynthesis of F0, precursor of the F420 cofactor, requires a unique two radical-SAM domain enzyme and tyrosine as substrate. J. Am. Chem. Soc. 134 (2012) 18173-18176. [PMID: 23072415]
2. Philmus, B., Decamps, L., Berteau, O. and Begley, T.P. Biosynthetic versatility and coordinated action of 5'-deoxyadenosyl radicals in deazaflavin biosynthesis. J. Am. Chem. Soc. 137 (2015) 5406-5413. [PMID: 25781338]
Accepted name: (R)-1-hydroxy-2-aminoethylphosphonate ammonia-lyase
Reaction: (1R)-(2-amino-1-hydroxyethyl)phosphonate = phosphonoacetaldehyde + NH3 (overall reaction)
(1a) (1R)-(2-amino-1-hydroxyethyl)phosphonate = (E)-2-aminoethenylphosphonate + H2O
(1b) (E)-2-aminoethenylphosphonate + H2O = phosphonoacetaldehyde + NH3
Glossary: phosphonoacetaldehyde = (2-oxoethyl)phosphonate
Other name(s): pbfA (gene name)
Systematic name: (1R)-(2-amino-1-hydroxyethyl)phosphonate ammonia-lyase
Comments: A pyridoxal-phosphate enzyme. This bacterial enzyme, characterized from the marine bacterium Vibrio splendidus, expands the substrate scope of a widespread pathway for the degradation of 2-aminoethylphosphonate. The enzyme is highly specific and does not act on the S enantiomer of 1-hydroxy-2-aminoethylphosphonate or other structurally related compounds.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Zangelmi, E., Stankovic, T., Malatesta, M., Acquotti, D., Pallitsch, K. and Peracchi, A. Discovery of a new, recurrent enzyme in bacterial phosphonate degradation: (R)-1-hydroxy-2-aminoethylphosphonate ammonia-lyase. Biochemistry 60 (2021) 1214-1225. [PMID: 33830741]
Accepted name: argininosuccinate lyase
Reaction: 2-(Nω-L-arginino)succinate = fumarate + L-arginine
For diagram click here.
Other name(s): arginosuccinase; argininosuccinic acid lyase; arginine-succinate lyase; N-(L-argininosuccinate) arginine-lyase; ω-N-(L-arginino)succinate arginine-lyase
Systematic name: 2-(Nω-L-arginino)succinate arginine-lyase (fumarate-forming)
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9027-34-3
References:
1. Davison, D.C. and Elliott, W.H. Enzymic reaction between arginine and fumarate in plant and animal tissue. Nature 169 (1952) 313-314. [PMID: 14910762]
Accepted name: adenylosuccinate lyase
Reaction: (1) N6-(1,2-dicarboxyethyl)AMP = fumarate + AMP
(2) (S)-2-[5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido]succinate = fumarate + 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
For diagram click here and another example click here.
Other name(s): adenylosuccinase; succino AMP-lyase; N6-(1,2-dicarboxyethyl)AMP AMP-lyase; 6-N-(1,2-dicarboxyethyl)AMP AMP-lyase
Systematic name: N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming)
Comments: Also acts on 1-(5-phosphoribosyl)-4-(N-succinocarboxamide)-5-aminoimidazole.
Links to other databases: BRENDA, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9027-81-0
References:
1. Carter, C.E. and Cohen, L.H. The preparation and properties of adenylosuccinase and adenylosuccinic acid. J. Biol. Chem. 222 (1956) 17-30. [PMID: 13366975]
Accepted name: ureidoglycolate lyase
Reaction: (S)-ureidoglycolate = glyoxylate + urea
For diagram of reaction click here.
Other name(s): ureidoglycolatase (ambiguous); ureidoglycolase (ambiguous); ureidoglycolate hydrolase (misleading); (S)-ureidoglycolate urea-lyase
Systematic name: (S)-ureidoglycolate urea-lyase (glyoxylate-forming)
Comments: This microbial enzyme is involved in the degradation of ureidoglycolate, an intermediate of purine degradation. Not to be confused with EC 3.5.1.116, ureidoglycolate amidohydrolase, which releases ammonia rather than urea.
Links to other databases: BRENDA, EXPASY, GTD, KEGG, MetaCyc, PDB, CAS registry number: 9014-57-7
References:
1. Trijbels, F. and Vogels, G.D. Allantoate and ureidoglycolate degradation by Pseudomonas aeruginosa. Biochim. Biophys. Acta 132 (1967) 115-126. [PMID: 6030341]
2. Werner, A.K., Romeis, T. and Witte, C.P. Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 6 (2010) 19-21. [PMID: 19935661]
Accepted name: purine imidazole-ring cyclase
Reaction: DNA 4,6-diamino-5-formamidopyrimidine = DNA adenine + H2O
Other name(s): DNA-4,6-diamino-5-formamidopyrimidine C8-N9-lyase (cyclizing); DNA-4,6-diamino-5-formamidopyrimidine 8-C,9-N-lyase (cyclizing)
Systematic name: DNA-4,6-diamino-5-formamidopyrimidine C8-N9-lyase (cyclizing; DNA-adenine-forming)
Comments: Also acts on 2,6-diamino-5-formamido-3,4-dihydro-4-oxopyrimidine residues. Brings about the reclosure of the imidazole rings of purine residues damaged by γ-rays.
Links to other databases: BRENDA, EXPASY, KEGG, CAS registry number: 95990-28-6
References:
1. Chetsanga, C.J. and Grigorian, C. In situ enzymatic reclosure of opened imidazole rings of purines in DNA damaged by γ-irradiation. Proc. Natl. Acad. Sci. USA 82 (1985) 633-637. [PMID: 3856219]
Accepted name: peptidylamidoglycolate lyase
Reaction: [peptide]-(2S)-2-hydroxyglycine = [peptide]-amide + glyoxylate
Other name(s): α-hydroxyglycine amidating dealkylase; peptidyl-α-hydroxyglycine α-amidating lyase; HGAD; PGL; PAL; peptidylamidoglycolate peptidylamide-lyase
Systematic name: [peptide]-(2S)-2-hydroxyglycine peptidyl-amide-lyase (glyoxylate-forming)
Comments: Requires zinc. The enzyme acts on the product of the reaction catalysed by EC 1.14.17.3 peptidylglycine monooxygenase, thus removing a terminal glycine residue and leaving a des-glycine peptide amide. In mammals, the two activities are part of a bifunctional protein.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number: 131689-50-4
References:
1. Katapodis, A.G., Ping, D. and May, S.W. A novel enzyme from bovine neurointermediate pituitary catalyzes dealkylation of α-hydroxyglycine derivatives, thereby functioning sequentially with peptidylglycine α-amidating monooxygenase in peptide amidation. Biochemistry 29 (1990) 6115-6120. [PMID: 2207061]
2. Bell, J., Ash, D.E., Snyder, L.M., Kulathila, R., Blackburn, N.J. and Merkler, D.J. Structural and functional investigations on the role of zinc in bifunctional rat peptidylglycine α-amidating enzyme. Biochemistry 36 (1997) 16239-16246. [PMID: 9405058]
Accepted name: γ-L-glutamyl-butirosin B γ-glutamyl cyclotransferase
Reaction: γ-L-glutamyl-butirosin B = butirosin B + 5-oxo-L-proline
Glossary: γ-L-glutamyl-butirosin B = (1R,2R,3S,4R,6S)-6-amino-4-{[(2R)-4-(γ-L-glutamylamino)-2-hydroxybutanoyl]amino}-3-hydroxy-2-(α-D-ribofuranosyloxy)cyclohexyl; γ-L-glutamyl-butirosin B γ-glutamyl cyclotransferase (5-oxoproline producing)
Other name(s): btrG (gene name)
Systematic name: γ-L-glutamyl-butirosin B γ-glutamyl cyclotransferase (5-oxo-L-proline producing)
Comments: The enzyme catalyses the last step in the biosynthesis of the aminoglycoside antibiotic butirosin B. The enzyme acts as a cyclotransferase, cleaving the amide bond via transamidation using the α-amine of the terminal γ-L-glutamate of the side chain, releasing it as the cyclic 5-oxo-L-proline.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Llewellyn, N.M., Li, Y. and Spencer, J.B. Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem. Biol. 14 (2007) 379-386. [PMID: 17462573]
Accepted name: glutathione-specific γ-glutamylcyclotransferase
Reaction: glutathione = L-cysteinylglycine + 5-oxo-L-proline
For diagram of reaction click here and mechanism click here
Other name(s): γ-GCG; CHAC (gene name); CHAC1 (gene name); CHAC2 (gene name)
Systematic name: glutathione γ-glutamyl cyclotransferase (5-oxo-L-proline producing)
Comments: The enzyme, found in bacteria, fungi and animals, is specific for glutathione (cf. EC 4.3.2.9, γ-glutamylcyclotransferase). The enzyme acts as a cyclotransferase, cleaving the amide bond via transamidation using the α-amine of the L-glutamyl residue, releasing it as the cyclic 5-oxo-L-proline.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Kumar, A., Tikoo, S., Maity, S., Sengupta, S., Sengupta, S., Kaur, A. and Bachhawat, A.K. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 13 (2012) 1095-1101. [PMID: 23070364]
2. Kaur, A., Gautam, R., Srivastava, R., Chandel, A., Kumar, A., Karthikeyan, S. and Bachhawat, A.K. ChaC2, an enzyme for slow turnover of cytosolic glutathione. J. Biol. Chem. 292 (2017) 638-651. [PMID: 27913623]
Accepted name: γ-glutamylamine cyclotransferase
Reaction: ε-(γ-L-glutamyl)-L-lysine = L-lysine + 5-oxo-L-proline
Other name(s): GGACT
Systematic name: ε-(γ-L-glutamyl)-L-lysine γ-glutamyl cyclotransferase (5-oxo-L-proline producing)
Comments: The enzyme, found in vertebrates, has no activity toward α-(γ-L-glutamyl)-L-amino acids (cf. EC 4.3.2.9, γ-glutamylcyclotransferase). The enzyme acts as a cyclotransferase, cleaving the amide bond via transamidation using the α-amine of the γ-L-glutamyl residue, releasing it as the cyclic 5-oxo-L-proline.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Fink, M.L., Chung, S.I. and Folk, J.E. γ-Glutamylamine cyclotransferase: specificity toward ε-(L-γ-glutamyl)-L-lysine and related compounds. Proc. Natl Acad. Sci. USA 77 (1980) 4564-4568. [PMID: 6107907]
2. Oakley, A.J., Coggan, M. and Board, P.G. Identification and characterization of γ-glutamylamine cyclotransferase, an enzyme responsible for γ-glutamyl-ε-lysine catabolism. J. Biol. Chem. 285 (2010) 9642-9648. [PMID: 20110353]
Accepted name: γ-glutamylcyclotransferase
Reaction: α-(γ-L-glutamyl)-L-amino acid = α-L-amino acid + 5-oxo-L-proline
Other name(s): γ-glutamyl-amino acid cyclotransferase; γ-L-glutamylcyclotransferase; L-glutamic cyclase; (5-L-glutamyl)-L-amino-acid 5-glutamyltransferase (cyclizing); GGCT
Systematic name: α-(γ-L-glutamyl)-L-amino-acid γ-glutamyl cyclotransferase (5-oxo-L-proline producing)
Comments: The enzyme, found in animals and plants, acts on derivatives of L-glutamate, L-2-aminobutanoate, L-alanine and glycine. The enzyme acts as a cyclotransferase, cleaving the amide bond via transamidation using the α-amine of the L-glutamyl residue, releasing it as the cyclic 5-oxo-L-proline.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Bodnaryk, R.P. and McGirr, L. Purification, properties and function of a unique γ-glutamyl cyclotransferase from the housefly, Musca domestica L. Biochim. Biophys. Acta 315 (1973) 352-362.
2. Orlowski, M., Richman, P.G. and Meister, A. Isolation and properties of γ-L-glutamylcyclotransferase from human brain. Biochemistry 8 (1969) 1048-1055. [PMID: 5781001]
3. Oakley, A.J., Yamada, T., Liu, D., Coggan, M., Clark, A.G. and Board, P.G. The identification and structural characterization of C7orf24 as γ-glutamyl cyclotransferase. An essential enzyme in the γ-glutamyl cycle. J. Biol. Chem. 283 (2008) 22031-22042. [PMID: 18515354]
4. Paulose, B., Chhikara, S., Coomey, J., Jung, H.I., Vatamaniuk, O. and Dhankher, O.P. A γ-glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell 25 (2013) 4580-4595. [PMID: 24214398]
Accepted name: imidazole glycerol-phosphate synthase
Reaction: (1) 5-[(5-phospho-1-deoxy-D-ribulos-1-ylamino)methylideneamino]-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide + L-glutamine = 5-amino-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide + D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate + L-glutamate (overall reaction)
(1a) L-glutamine + H2O = L-glutamate + NH3
(1b) 5-[(5-phospho-1-deoxy-D-ribulos-1-ylamino)methylideneamino]-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide + NH3 = 5-amino-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide + D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate + H2O
For diagram of reaction click here.
Other name(s): IGP synthase; hisFH (gene names); HIS7 (gene name)
Systematic name: 5-[(5-phospho-1-deoxy-D-ribulos-1-ylamino)methylideneamino]-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate-lyase (L-glutamine-hydrolysing; 5-amino-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamide-forming)
Comments: The enzyme is involved in histidine biosynthesis, as well as purine nucleotide biosynthesis. The enzymes from archaea and bacteria are heterodimeric. A glutaminase component (cf. EC 3.5.1.2, glutaminase) produces an ammonia molecule that is transferred by a 25 Å tunnel to a cyclase component, which adds it to the imidazole ring, leading to lysis of the molecule and cyclization of one of the products. The glutminase subunit is only active within the dimeric complex. In fungi and plants the two subunits are combined into a single polypeptide.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Klem, T.J. and Davisson, V.J. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry 32 (1993) 5177-5186. [PMID: 8494895]
2. Fujimori, K. and Ohta, D. An Arabidopsis cDNA encoding a bifunctional glutamine amidotransferase/cyclase suppresses the histidine auxotrophy of a Saccharomyces cerevisiae his7 mutant. FEBS Lett. 428 (1998) 229-234. [PMID: 9654139]
3. Beismann-Driemeyer, S. and Sterner, R. Imidazole glycerol phosphate synthase from Thermotoga maritima. Quaternary structure, steady-state kinetics, and reaction mechanism of the bienzyme complex. J. Biol. Chem 276 (2001) 20387-20396. [PMID: 11264293]
4. Douangamath, A., Walker, M., Beismann-Driemeyer, S., Vega-Fernandez, M.C., Sterner, R. and Wilmanns, M. Structural evidence for ammonia tunneling across the (β α)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure 10 (2002) 185-193. [PMID: 11839304]
5. Chaudhuri, B.N., Lange, S.C., Myers, R.S., Davisson, V.J. and Smith, J.L. Toward understanding the mechanism of the complex cyclization reaction catalyzed by imidazole glycerolphosphate synthase: crystal structures of a ternary complex and the free enzyme. Biochemistry 42 (2003) 7003-7012. [PMID: 12795595]
Accepted name: (3R)-3-[(carboxylmethyl)amino]fatty acid synthase
Reaction: (3R)-3-[(carboxylmethyl)amino]fatty acid + an [acyl-carrier protein] = a (2E)-unsaturated fatty acyl-[acyl-carrier protein] + glycine + H2O
Other name(s): scoD (gene name); mmaD (gene name)
Systematic name: (3R)-3-[(carboxylmethyl)amino]fatty acid glycine-lyase ((2E)-unsaturated fatty acyl-[acyl-carrier protein]-forming)
Comments: The enzyme, found in some actinobacterial species, participates in the biosynthesis of isonitrile-containing lipopeptides. It catalyses the formation of (3R)-3-[(carboxylmethyl)amino]fatty acid by the addition of glycine and the release of the product from the acyl-carrier protein.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Harris, N.C., Sato, M., Herman, N.A., Twigg, F., Cai, W., Liu, J., Zhu, X., Downey, J., Khalaf, R., Martin, J., Koshino, H. and Zhang, W. Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria. Proc. Natl. Acad. Sci. USA 114 (2017) 7025-7030. [PMID: 28634299]
2. Harris, N.C., Born, D.A., Cai, W., Huang, Y., Martin, J., Khalaf, R., Drennan, C.L. and Zhang, W. Isonitrile formation by a non-heme iron(II)-dependent oxidase/decarboxylase. Angew. Chem. Int. Ed. Engl. 57 (2018) 9707-9710. [PMID: 29906336]
Accepted name: 3-ketovalidoxylamine C-N-lyase
Reaction: 4-nitrophenyl-3-ketovalidamine = 4-nitroaniline + 5-D-(5/6)-5-C-(hydroxymethyl)-2,6-dihydroxycyclohex-2-en-1-one
Other name(s): 3-ketovalidoxylamine A C-N-lyase; p-nitrophenyl-3-ketovalidamine p-nitroaniline lyase; 4-nitrophenyl-3-ketovalidamine 4-nitroaniline-lyase
Systematic name: 4-nitrophenyl-3-ketovalidamine 4-nitroaniline-lyase [5-D-(5/6)-5-C-(hydroxymethyl)-2,6-dihydroxycyclohex-2-en-1-one-forming]
Comments: Requires Ca2+. Eliminates 4-nitroaniline from 4-nitrophenyl-3-ketovalidamine, or 4-nitrophenol from 4-nitrophenyl-α-D-3-dehydroglucoside. Involved in the degradation of the fungicide validamycin A by Flavobacterium saccharophilum.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 99889-98-2
References:
1. Asano, N., Takeuchi, M., Ninomiya, K., Kameda, Y. and Matsui, K. Microbial degradation of validamycin A by Flavobacterium saccharophilum. Enzymatic cleavage of C-N linkage in validoxylamine A. J. Antibiot. 37 (1984) 859-867. [PMID: 6548220]
2. Takeuchi, M., Asano, N., Kameda, Y. and Matsui, K. Purification and properties of 3-ketovalidoxylamine A C-N lyase from Flavobacterium saccharophilum. J. Biochem. (Tokyo) 98 (1985) 1631-1638. [PMID: 4093450]
[EC 4.3.3.2 Transferred entry: strictosidine synthase. Now EC 3.5.99.13, strictosidine synthase (EC 4.3.3.2 created 1990, deleted 2024)]
[EC 4.3.3.3 Transferred entry: deacetylisoipecoside synthase. Now EC 3.5.99.15, deacetylisoipecoside synthase (EC 4.3.3.3 created 2000, deleted 2024)]
[EC 4.3.3.4 Transferred entry: deacetylipecoside synthase. Now EC 3.5.99.16, deacetylipecoside synthase (EC 4.3.3.4 created 2000, deleted 2024)]
Accepted name: 4'-demethylrebeccamycin synthase
Reaction: 4'-O-demethylrebeccamycin + H2O = dichloro-arcyriaflavin A + β-D-glucose
For diagram of reaction click here
Glossary: dichloro-arcyriaflavin A = rebeccamycin aglycone
Other name(s): arcyriaflavin A N-glycosyltransferase; RebG
Systematic name: 4'-demethylrebeccamycin D-glucose-lyase
Comments: This enzyme catalyses a step in the biosynthesis of rebeccamycin, an indolocarbazole alkaloid produced by the bacterium Lechevalieria aerocolonigenes. The enzyme is a glycosylase, and acts in the reverse direction to that shown. It has a wide substrate range, and was shown to glycosylate several substrates, including the staurosporine aglycone, EJG-III-108A, J-104303, 6-N-methyl-arcyriaflavin C and indolo-[2,3-a]-carbazole [1,2].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Ohuchi, T., Ikeda-Araki, A., Watanabe-Sakamoto, A., Kojiri, K., Nagashima, M., Okanishi, M. and Suda, H. Cloning and expression of a gene encoding N-glycosyltransferase (ngt) from Saccharothrix aerocolonigenes ATCC39243. J. Antibiot. (Tokyo) 53 (2000) 393-403. [PMID: 10866221]
2. Zhang, C., Albermann, C., Fu, X., Peters, N.R., Chisholm, J.D., Zhang, G., Gilbert, E.J., Wang, P.G., Van Vranken, D.L. and Thorson, J.S. RebG- and RebM-catalyzed indolocarbazole diversification. Chembiochem 7 (2006) 795-804. [PMID: 16575939]
Accepted name: pyridoxal 5'-phosphate synthase (glutamine hydrolysing)
Reaction: D-ribose 5-phosphate + D-glyceraldehyde 3-phosphate + L-glutamine = pyridoxal 5'-phosphate + L-glutamate + 3 H2O + phosphate (overall reaction)
(1a) L-glutamine + H2O = L-glutamate + NH3
(1b) D-ribose 5-phosphate + D-glyceraldehyde 3-phosphate + NH3 = pyridoxal 5'-phosphate + 4 H2O + phosphate
Other name(s): PdxST; pyridoxal 5'-phosphate synthase (glutamine hydrolyzing)
Systematic name: D-ribose 5-phosphate,D-glyceraldehyde 3-phosphate pyridoxal 5'-phosphate-lyase
Comments: The ammonia is provided by the glutaminase subunit and channeled to the active site of the lyase subunit by a 100 Å tunnel. The enzyme can also use ribulose 5-phosphate and dihydroxyacetone phosphate. The enzyme complex is found in aerobic bacteria, archaea, fungi and plants.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Burns, K.E., Xiang, Y., Kinsland, C.L., McLafferty, F.W. and Begley, T.P. Reconstitution and biochemical characterization of a new pyridoxal-5'-phosphate biosynthetic pathway. J. Am. Chem. Soc. 127 (2005) 3682-3683. [PMID: 15771487]
2. Raschle, T., Amrhein, N. and Fitzpatrick, T.B. On the two components of pyridoxal 5'-phosphate synthase from Bacillus subtilis. J. Biol. Chem. 280 (2005) 32291-32300. [PMID: 16030023]
3. Strohmeier, M., Raschle, T., Mazurkiewicz, J., Rippe, K., Sinning, I., Fitzpatrick, T.B. and Tews, I. Structure of a bacterial pyridoxal 5'-phosphate synthase complex. Proc. Natl. Acad. Sci. USA 103 (2006) 19284-19289. [PMID: 17159152]
4. Raschle, T., Arigoni, D., Brunisholz, R., Rechsteiner, H., Amrhein, N. and Fitzpatrick, T.B. Reaction mechanism of pyridoxal 5'-phosphate synthase. Detection of an enzyme-bound chromophoric intermediate. J. Biol. Chem. 282 (2007) 6098-6105. [PMID: 17189272]
5. Hanes, J.W., Keresztes, I. and Begley, T.P. Trapping of a chromophoric intermediate in the Pdx1-catalyzed biosynthesis of pyridoxal 5'-phosphate. Angew. Chem. Int. Ed. Engl. 47 (2008) 2102-2105. [PMID: 18260082]
6. Hanes, J.W., Burns, K.E., Hilmey, D.G., Chatterjee, A., Dorrestein, P.C. and Begley, T.P. Mechanistic studies on pyridoxal phosphate synthase: the reaction pathway leading to a chromophoric intermediate. J. Am. Chem. Soc. 130 (2008) 3043-3052. [PMID: 18271580]
7. Hanes, J.W., Keresztes, I. and Begley, T.P. 13C NMR snapshots of the complex reaction coordinate of pyridoxal phosphate synthase. Nat. Chem. Biol. 4 (2008) 425-430. [PMID: 18516049]
8. Wallner, S., Neuwirth, M., Flicker, K., Tews, I. and Macheroux, P. Dissection of contributions from invariant amino acids to complex formation and catalysis in the heteromeric pyridoxal 5-phosphate synthase complex from Bacillus subtilis. Biochemistry 48 (2009) 1928-1935. [PMID: 19152323]
Accepted name: 4-hydroxy-tetrahydrodipicolinate synthase
Reaction: pyruvate + L-aspartate-4-semialdehyde = (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate + H2O
For diagram of reaction click here
Glossary: (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate = (2S,4S)-4-hydroxy-2,3,4,5-tetrahydropyridine-2,6-dicarboxylate
Other name(s): dihydrodipicolinate synthase (incorrect); dihydropicolinate synthetase (incorrect); dihydrodipicolinic acid synthase (incorrect); L-aspartate-4-semialdehyde hydro-lyase (adding pyruvate and cyclizing); dapA (gene name).
Systematic name: L-aspartate-4-semialdehyde hydro-lyase [adding pyruvate and cyclizing; (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinate-forming]
Comments: The reaction can be divided into three consecutive steps: Schiff base formation with pyruvate, the addition of L-aspartate-semialdehyde, and finally transimination leading to cyclization with simultaneous dissociation of the product. The product of the enzyme was initially thought to be (S)-2,3-dihydrodipicolinate [1,2], and the enzyme was classified accordingly as EC 4.2.1.52, dihydrodipicolinate synthase. Later studies of the enzyme from the bacterium Escherichia coli have suggested that the actual product of the enzyme is (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate [3], and thus the enzyme has been reclassified as 4-hydroxy-tetrahydrodipicolinate synthase. However, the identity of the product is still controversial, as more recently it has been suggested that it may be (S)-2,3-dihydrodipicolinate after all [5].
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Yugari, Y. and Gilvarg, C. The condensation step in diaminopimelate synthesis. J. Biol. Chem. 240 (1965) 4710-4716. [PMID: 5321309]
2. Blickling, S., Renner, C., Laber, B., Pohlenz, H.D., Holak, T.A. and Huber, R. Reaction mechanism of Escherichia coli dihydrodipicolinate synthase investigated by X-ray crystallography and NMR spectroscopy. Biochemistry 36 (1997) 24-33. [PMID: 8993314]
3. Devenish, S.R., Blunt, J.W. and Gerrard, J.A. NMR studies uncover alternate substrates for dihydrodipicolinate synthase and suggest that dihydrodipicolinate reductase is also a dehydratase. J. Med. Chem. 53 (2010) 4808-4812. [PMID: 20503968]
4. Soares da Costa, T.P., Muscroft-Taylor, A.C., Dobson, R.C., Devenish, S.R., Jameson, G.B. and Gerrard, J.A. How essential is the ’essential’ active-site lysine in dihydrodipicolinate synthase. Biochimie 92 (2010) 837-845. [PMID: 20353808]
5. Karsten, W.E., Nimmo, S.A., Liu, J. and Chooback, L. Identification of 2,3-dihydrodipicolinate as the product of the dihydrodipicolinate synthase reaction from Escherichia coli. Arch. Biochem. Biophys. 653 (2018) 50-62. [PMID: 29944868]
Accepted name: mimosinase
Reaction: L-mimosine + H2O = 3-hydroxy-4H-pyrid-4-one + pyruvate + NH3 (overall reaction)
(1a) L-mimosine = 3-hydroxy-4H-pyrid-4-one + 2-aminoprop-2-enoate
(1b) 2-aminoprop-2-enoate = 2-iminopropanoate (spontaneous)
(1c) 2-iminopropanoate + H2O = pyruvate + NH3 (spontaneous)
Glossary: L-mimosine = (2S)-2-amino-3-[3-hydroxy-4-oxopyridin-1(4H)-yl]propanoate
Other name(s): mimosine amidohydrolase (incorrect)
Systematic name: (2S)-2-amino-3-[3-hydroxy-4-oxopyridin-1(4H)-yl]propanoate 3-hydroxy-4H-pyrid-4-one-lyase (2-aminoprop-2-enoate-forming)
Comments: A pyridoxal-phosphate protein. The enzyme degrades the toxic amino acid L-mimosine. It cleaves a carbon-nitrogen bond, releasing 3-hydroxy-4H-pyrid-4-one and an unstable enamine product that tautomerizes to an imine form, which undergoes a hydrolytic deamination to form pyruvate and ammonia. It is thought to have evolved from EC 4.4.1.13, cysteine-S-conjugate β-lyase. It has been described in both mimosine-producing plants and some bacteria.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Tangendjaja, B., Lowry, J.B. and Wills, R.H. Isolation of a mimosine degrading enzyme from Leucaena leaf. J. Sci. Food Agric. 37 (1986) 523-526.
2. Negi, V.S., Bingham, J.P., Li, Q.X. and Borthakur, D. A carbon-nitrogen lyase from Leucaena leucocephala catalyzes the first step of mimosine degradation. Plant Physiol. 164 (2014) 922-934. [PMID: 24351687]
3. Oogai, S., Fukuta, M., Watanabe, K., Inafuku, M. and Oku, H. Molecular characterization of mimosinase and cystathionine β-lyase in the Mimosoideae subfamily member Mimosa pudica. J. Plant Res. 132 (2019) 667-680. [PMID: 31368041]
4. Oogai, S., Fukuta, M., Inafuku, M. and Oku, H. Isolation and characterization of mimosine degrading enzyme from Arthrobacter sp. Ryudai-S1. World J. Microbiol. Biotechnol. 38 (2022) 172. [PMID: 35908235]
Accepted name: indigoidine synthase
Reaction: 2 ATP + 2 L-glutamine + O2 + 2 FMN = 2 AMP + 2 diphosphate + indigoidine + 2 H2O + 2 FMNH2 (overall reaction)
(1) 2 ATP + 2 L-glutamine + 2 FMN = 2 AMP + 2 diphosphate + 2 3-amino-1,5-dihydropyridine-2,6-dione + FMNH2
(2) 2 3-amino-1,5-dihydropyridine-2,6-dione + O2 = indigoidine + 2 H2O (spontaneous)
For diagram of reaction, click here
Glossary: indigoidine = 3-(5-amino-2-hydroxy-6-oxo-1H-pyridin-3-yl)-5-iminopyridine-2,6-dione
Other name(s): bspA (gene name)
Systematic name: L-glutamine oxidoreductase/cyclase (3-amino-1,5-dihydropyridine-2,6-dione-forming)
Comments: The enzyme, found in a number of bacterial strains, is a non-ribosomal peptide synthase (NRPS). The enzyme forms 3-amino-1,5-dihydropyridine-2,6-dione, which undergoes spontaneous oxidation to form the blue pigment indigoidine.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Reverchon, S., Rouanet, C., Expert, D. and Nasser, W. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184 (2002) 654-665. [PMID: 11790734]
2. Takahashi, H., Kumagai, T., Kitani, K., Mori, M., Matoba, Y. and Sugiyama, M. Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J. Biol. Chem. 282 (2007) 9073-9081. [PMID: 17237222]
3. Walsh, C.T. and Wencewicz, T.A. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 30 (2013) 175-200. [PMID: 23051833]
[EC 4.3.99.2 Transferred entry: carboxybiotin decarboxylase. Now EC 7.2.4.1, carboxybiotin decarboxylase (EC 4.3.99.2 created 2008, deleted 2018)]
Accepted name: 7-carboxy-7-deazaguanine synthase
Reaction: 6-carboxy-5,6,7,8-tetrahydropterin = 7-carboxy-7-carbaguanine + NH3
For diagram of reaction click here.
Glossary: 7-carboxy-7-carbaguanine = 7-carboxy-7-deazaguanine
Other name(s): 7-carboxy-7-carbaguanine synthase; queE (gene name)
Systematic name: 6-carboxy-5,6,7,8-tetrahydropterin ammonia-lyase
Comments: Requires Mg2+. The enzyme is a member of the superfamily of S-adenosyl-L-methionine-dependent radical (radical AdoMet) enzymes. Binds a [4Fe-4S] cluster that is coordinated by 3 cysteines and an exchangeable S-adenosyl-L-methionine molecule. The S-adenosyl-L-methionine is catalytic as it is regenerated at the end of the reaction. The reaction is part of the biosynthesis pathway of queuosine.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. McCarty, R.M., Somogyi, A., Lin, G., Jacobsen, N.E. and Bandarian, V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine 5'-triphosphate in four steps. Biochemistry 48 (2009) 3847-3852. [PMID: 19354300]
2. McCarty, R.M., Krebs, C. and Bandarian, V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry 52 (2013) 188-198. [PMID: 23194065]
Accepted name: choline trimethylamine-lyase
Reaction: choline = trimethylamine + acetaldehyde
Other name(s): cutC (gene name)
Systematic name: choline trimethylamine-lyase (acetaldehyde-forming)
Comments: The enzyme utilizes a glycine radical to break the C-N bond in choline. Found in choline-degrading anaerobic bacteria.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Craciun, S. and Balskus, E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 109 (2012) 21307-21312. [PMID: 23151509]
Accepted name: nitrosuccinate lyase
Reaction: 2-nitrobutanedioate = fumarate + nitrite
Glossary: 2-nitrobutanedioate = nitrosuccinate
Other name(s): creD (gene name)
Systematic name: 2-nitrobutanedioate lyase (fumarate-forming)
Comments: The enzyme, found in some Actinobacteria, is involved in a pathway that forms nitrite, which is subsequently used to generate a diazo group in some secondary metabolites.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Sugai, Y., Katsuyama, Y. and Ohnishi, Y. A nitrous acid biosynthetic pathway for diazo group formation in bacteria. Nat. Chem. Biol. 12 (2016) 73-75. [PMID: 26689788]
2. Hagihara, R., Katsuyama, Y., Sugai, Y., Onaka, H. and Ohnishi, Y. Novel desferrioxamine derivatives synthesized using the secondary metabolism-specific nitrous acid biosynthetic pathway in Streptomyces davawensis, J. Antibiot. (Tokyo) 71 (2018) 911-919. [PMID: 30120394]