An asterisk before 'EC' indicates that this is an amendment to an existing enzyme rather than a new enzyme entry.
Common name: 1-deoxy-D-xylulose-5-phosphate reductoisomerase
Reaction: 2-C-methyl-D-erythritol 4-phosphate + NADP+ = 1-deoxy-D-xylulose 5-phosphate + NADPH + H+
For diagram click here.
Other name(s): DXP-reductoisomerase
Systematic name: 2-C-methyl-D-erythritol-4-phosphate:NADP+ oxidoreductase (isomerizing)
Comments: The enzyme requires Mn2+, Co2+ or Mg2+ for activity, with the first being most effective. The enzyme from several eubacteria, including E. coli, forms part of an alternative nonmevalonate pathway for terpenoid biosynthesis.
References:
1. Takahashi, S., Kuzuyama, T., Watanabe, H. and Seto, H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95 (1998) 9879-9884. [PMID: 9707569]
Common name: D-mannitol oxidase
Reaction: mannitol + O2 = mannose + H2O2
Other name(s): mannitol oxidase; D-arabinitol oxidase
Systematic name: mannitol:oxygen oxidoreductase (cyclizing)
Comments: Also catalyses the oxidation of D-arabinitol and, to a lesser extent, D-glucitol (sorbitol), whereas L-arabinitol is not a good substrate. The enzyme from the snails Helix aspersa and Arion ater is found in a specialised tubular organelle that has been termed the mannosome.
References:
1. Vorhaben, J.E., Scott, J.F., Smith, D.D. and Campbell, J.W. Mannitol oxidase: partial purification and characterisation of the membrane-bound enzyme from the snail Helix aspersa. Int. J. Biochem. 18 (1986) 337-344. [PMID: 3519307]
2. Large, A.T., Jones, C.J.P. and Connock, M.J. The association of mannitol oxidase with a distinct organelle in the digestive gland of the terrestrial slug Arion ater. Protoplasma 175 (1993) 93-101.
Common name: nitrite reductase (cytochrome; NO-forming)
Reaction: nitric oxide + H2O + ferricytochrome c = nitrite + ferrocytochrome c + 2 H+
Other name(s): [nitrite reductase (cytochrome)]
Systematic name: nitric-oxide:ferricytochrome-c oxidoreductase
Comments: A copper protein. Cytochrome c-552 or cytochrome c-553 from Pseudomonas denitrificans acts as acceptor.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 37256-41-0
References:
1. Miyata, M. and Mori, T. Studies on denitrification. X. The "denitrifying enzyme" as a nitrite reductase and the electron donating system for denitrification. J. Biochem. (Tokyo) 66 (1969) 463-471. [PMID: 5354021]
Common name: nitrite reductase (cytochrome; ammonia-forming)
Reaction: ammonia + 2 H2O + 6 ferricytochrome c = nitrite + 6 ferrocytochrome c + 7 H+
Other names: cytochrome c nitrite reductase; multiheme nitrite reductase
Systematic name: ammonia:ferricytochrome-c oxidoreductase
Comments: Found as a multiheme cytochrome in many bacteria. The enzyme from E. coli contains five hemes c and requires Ca2+. It also reduces nitric oxide and hydroxylamine to ammonia, and sulfite to sulfide.
References:
1. Einsle, O., Messerschmidt, A., Stach, P. Bourenkov, G.P., Bartunik, H.D., Huber, R. and Kroneck, P.M.H. Structure of cytochrome c nitrite reductase. Nature 400 (1999) 476-480. [PMID: 10440380]
Common name: 4-hydroxymandelate synthase
Reaction: 4-hydroxyphenylpyruvate + O2 = 4-hydroxymandelate + CO2
Other name(s): 4-hydroxyphenylpyruvate dioxygenase II
Systematic name: 4-hydroxyphenylpyruvate:oxygen oxidoreductase (decarboxylating)
Comments: Requires Fe2+. Involved in the biosynthesis of the vancomycin group of glycopeptide antibiotics.
References:
1. Choroba, O.W., Williams, D.H. and Spencer, J.B. Biosynthesis of the vancomycin group of antibiotics: involvement of an unusual dioxygenase in the pathway to (S)-4-hydroxyphenylglycine. J. Am. Chem. Soc. 122 (2000) 5389-5390.
*EC 1.18. Acting on Iron-Sulfur Proteins as Donors
Common name: rubredoxin-NAD+ reductase
Reaction: reduced rubredoxin + NAD = oxidized rubredoxin + NADH + H+
Glossary entries:
rubredoxin
Other name(s): rubredoxin reductase; rubredoxin-nicotinamide adenine dinucleotide reductase; dihydronicotinamide adenine dinucleotide-rubredoxin reductase; reduced nicotinamide adenine dinucleotide-rubredoxin reductase; NADH-rubredoxin reductase; rubredoxin-NAD reductase; NADH: rubredoxin oxidoreductase; DPNH-rubredoxin reductase; NADH-rubredoxin oxidoreductase
Systematic name: rubredoxin:NAD+ oxidoreductase
Comments: Requires FAD. The enzyme from Clostridium acetobutylicum reduces rubredoxin, ferricyanide and dichlorophenolindophenol, but not ferredoxin or flavodoxin. The reaction does not occur when NADPH is substituted for NADH. Contains iron at the redox centre. Formerly EC 1.6.7.2.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 9032-27-3
References:
1. Peterson, J.A., Kusunose, M., Kusunose, E. and Coon, M.J. Enzymatic ω-oxidation. II. Function of rubredoxin as the electron carrier in ω-hydroxylation. J. Biol. Chem. 242 (1967) 4334-4340. [PMID: 4294330]
2. Ueda, T., Lode, E.T. and Coon, M.J. Enzymatic ω-oxidation. VI. Isolation of homogeneous reduced diphosphopyridine nucleotide-rubredoxin reductase. J. Biol. Chem. 247 (1972) 2109-2116. [PMID: 4335861]
3. Ueda, T., Lode, E.T. and Coon, M.J. Enzymatic oxidation. VII. Reduced diphosphopyridine nucleotide-rubredoxin reductase: properties and function as an electron carrier in hydroxylation. J. Biol. Chem. 247 (1972) 5010-5016. [PMID: 4403503]
4. Petitdemange, H., Marczak, R., Blusson, H. and Gay, R. Isolation and properties of reduced nicotinamide adenine dinucleotide rubredoxin oxidoreductase of Clostridium acetobutylicum. Biochem. Biophys. Res. Commun. 91 (1979) 1258-1265. [PMID: 526302]
Common name: rubredoxin-NAD(P)+ reductase
Reaction: reduced rubredoxin + NAD(P)+ = oxidized rubredoxin + NAD(P)H + H+
Glossary entries:
benzyl viologen = 1,1'-dibenzyl-4,4'-bipyridinium
2,6-dichloroindophenol = 4-(2,6-dichloro-4-hydroxyphenylimino)cyclohexa-2,5-dien-1-one
menadione = 2-methyl-1,4-naphthoquinone
rubredoxin
Other name(s): rubredoxin-nicotinamide adenine dinucleotide (phosphate) reductase; rubredoxin-nicotinamide adenine; dinucleotide phosphate reductase; NAD(P)-rubredoxin oxidoreductase; NAD(P)H-rubredoxin oxidoreductase
Systematic name: rubredoxin:NAD(P) oxidoreductase
Comments: The enzyme from Pyrococcus furiosis requires FAD. It reduces a number of electron carriers, including benzyl viologen, menadione and 2,6-dichloroindophenol, but rubredoxin is the most efficient. Ferredoxin is not utilized.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number:
References:
1. Petitdemange, H., Blusson, H. and Gay, R. Detection of NAD(P)H-rubredoxin oxidoreductases in Clostridia. Anal. Biochem. 116 (1981) 564-570. [PMID: 6274224]
2. Ma, K. and Adams, M.W.W. A hyperactive NAD(P)H:rubredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 181 (1999) 5530-5533. [PMID: 10464233]
EC 1.18.96 With other, known, acceptors
Common name: superoxide reductase
Reaction: reduced rubredoxin + superoxide + 2 H+ = H2O2 + rubredoxin
Glossary entries:
rubredoxin
superoxide = O2•-
Other name(s): neelaredoxin; desulfoferrodoxin
Systematic name: rubredoxin:superoxide oxidoreductase
Comments: The enzyme contains non-heme iron.
References:
1. Jenney Jr., F.E., Verhagen, M.F.J.M., Cui, X. and Adams, M.W.W. Anaerobic microbes: Oxygen detoxification without superoxide dismutase. Science 286 (1999) 306-309. [PMID: 10514376]
2. Yeh, A.P., Hu, Y., Jenney Jr., F.E., Adams, M.W.W. and Rees, D.C. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry 39 (2000) 2499-2508. [PMID: 10704199]
3. Lombard, M., Fontecave, M., Touati, D. and Niviere, V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J. Biol. Chem. 275 (2000) 115-121. [PMID: 10617593]
4. Abreu, I.A.,. Saraiva, L.M., Carita, J., Huber, H., Stetter, K.O., Cabelli, D.and Teixeira, M. Oxygen detoxification in the strict anaerobic archaeon Archaeoglobus fulgidus: superoxide scavenging by neelaredoxin. Mol. Microbiol. 38 (2000) 322-334. [PMID: 11069658]
Common name: tetrachloroethene reductive dehalogenase
Reaction: trichloroethene + chloride + acceptor = tetrachloroethene + reduced acceptor
Glossary entries:
methyl viologen = 1,1'-dimethyl-4,4'-bipyridinium
Other name(s): tetrachloroethene reductase
Systematic name: acceptor:trichloroethene oxidoreductase (chlorinating)
Comments: This enzyme allows the common pollutant tetrachloroethene to support bacterial growth and is responsible for disposal of a number of chlorinated hydrocarbons by this organism. The reaction occurs in the reverse direction. The enzyme also reduces trichloroethene to dichloroethene. Although the physiological reductant is unknown, the supply of reductant in some organisms is via reduced menaquinone, itself formed from molecular hydrogen, via EC 1.12.99.3 (hydrogen:quinone oxidoreductase). The enzyme contains a corrinoid and two iron-sulfur clusters. Methyl viologen can act as electron donor.
References:
1. Holliger, C, Wohlfarth, G. and Diekert, G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22 (1998/1999) 383-398.
2. Glod, G., Angst, W., Holliger, C. and Schwarzenbach, R.P. Corrinoid-mediated reduction of tetrachloroethene, trichloroethene, and trichlorofluoroethene in homogeneous aqueous solution: Reaction kinetics and reaction mechanisms. Environ. Sci. Technol. 31 (1997) 253-260.
3. Neumann, A., Wohlfarth, G. and Diekert, G. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271 (1996) 16515-16519. [PMID: 8663199]
4. Schumacher, W., Holliger, C., Zehnder, A.J.B. and Hagen, W.R. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409 (1997) 421-425. [PMID: 9224702]
5. Schumacher, W. and Holliger, C. The proton/electron ratio of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethene in "Dehalobacter restrictus". J. Bacteriol. 178 (1996) 2328-2333. [PMID: 8636034]
Common name: jasmonate O-methyltransferase
Reaction: S-adenosyl-L-methionine + jasmonate = S-adenosyl-L-homocysteine + methyljasmonate
Glossary entries:
jasmonic acid = {(1R,2R)-3-oxo-2-[(Z)pent-2-enyl]cyclopent-2-enyl}acetic acid.
Other name(s): jasmonic acid carboxyl methyltransferase
Systematic name: S-adenosyl-L-methionine:jasmonate O-methyltransferase
Comments: 9,10-Dihydrojasmonic acid is a poor substrate for the enzyme. The enzyme does not convert 12-oxo-phytodienoic acid (a precursor of jasmonic acid), salicylic acid, benzoic acid, linolenic acid or cinnamic acid into their corresponding methyl esters. Enzyme activity is inhibited by the presence of divalent cations, e.g., Ca2+, Cu2+, Mg2+ and Zn2+.
References:
1. Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S. and Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98 (2001) 4788-4793. [PMID: 11287667]
Common name: phospholipid:diacylglycerol acyltransferase
Reaction: phospholipid + 1,2-diacylglycerol = lysophospholipid + triacylglycerol
Glossary entries:
ricinoleic acid = (9Z,12R)-12-hydroxyoctadec-9-enoic acid
vernolic acid = (9Z,12S,13R)-12,13-epoxyoctadec-9-enoic acid.
Other name(s): PDAT
Systematic name: phospholipid:1,2-diacyl-sn-glycerol O-acyltransferase
Comments: This enzyme differs from EC 2.3.1.20, diacylglycerol O-acyltransferase, by synthesising triacylglycerol using an acyl-CoA-independent mechanism. The specificity of the enzyme for the acyl group in the phospholipid varies with species, e.g., the enzyme from castor bean (Ricinus communis) preferentially incorporates vernoloyl (12,13-epoxyoctadec-9-enoyl) groups into triacylglycerol, whereas that from the hawk's beard (Crepis palaestina) incorporates both ricinoleoyl (12-hydroxyoctadec-9-enoyl) and vernoloyl groups. The enzyme from the yeast Saccharomyces cerevisiae specifically transfers acyl groups from the sn-2 position of the phospholipid to diacylglycerol, thus forming an sn-1-lysophospholipid.
References:
1. Dahlqvist, A., Stähl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., Ronne, H. and Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97 (2000) 6487-6492. [PMID: 10829075]
Common name: hyaluronan synthase
Reaction: n UDP-N-acetyl-D-glucosamine + n UDP-D-glucuronate = [β-N-acetyl-D-glucosaminyl(1 4)β-D-glucuronosyl(1
3)]n+ 2n UDP
Comments: The enzyme from Streptococcus Group A and Group C requires Mg2+. It is highly specific for UDP-GlcNAc and UDP-GlcA; no copolymerization is observed if either is replaced by UDP-Glc, UDP-Gal, UDP-GalNAc or UDP-GalA. Similar enzymes have been found in a variety of organisms.
References:
1. DeAngelis, P.L., Papaconstantinou, J. and Weigel, P.H. Molecular cloning, identification and sequence of the hyaluronan synthase gene from Group A Streptococcus pyogenes. J. Biol. Chem. 268 (1993) 19181-19184. [PMID: 8366070]
2. Jing, W. and DeAngelis, P.L. Dissection of the two transferase activities of the Pasteurella multocida hyaluronan synthase: two active sites exist in one polypeptide. Glycobiology 10 (2000) 883-889. [PMID: 10988250]
3. DeAngelis, P.L. Molecular directionality of polysaccharide polymerization by the Pasteurella multocida hyaluronan synthase. J. Biol. Chem. 274 (1993) 26557-26562. [PMID: 10473619]
4. Buckeridge, M.S., Vergara, C.E., Carpita, N.C. The mechanism of synthesis of a mixed-linkage (1 3),(1
4)β-D-glucan in maize. Evidence for multiple sites of glucosyl transfer in the synthase complex. Plant Physiol. 120 (1999) 1105-1116.
Common name: glucosylglycerol-phosphate synthase
Reaction: ADPglucose + sn-glycerol 3-phosphate = 2-(β-D-glucosyl)-sn-glycerol 3-phosphate + ADP
Systematic name: ADPglucose:sn-glycerol-3-phosphate 2-β-D-glucosyltransferase
Comments: Acts with EC 3.1.3.69 (glucosylglycerol phosphatase) to form glucosylglycerol, an osmolyte that endows cyanobacteria with resistance to salt.
References:
1. Hagemann, M. and Erdmann, N. Activation and pathway of glucosylglycerol biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 140 (1994) 1427-1431.
2. Marin, K., Zuther, E., Kerstan, T., Kunert, A. and Hagemann, M. The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosylglycerol-phosphate synthase is involved in osmolyte synthesis. J. Bacteriol. 180 (1998) 4843-4849. [PMID: 9733686]
Common name: ADP-specific phosphofructokinase
Reaction: ADP + D-fructose 6-phosphate = AMP + D-fructose 1,6-bisphosphate
Other name(s): ADP-6-phosphofructokinase, ADP-dependent phosphofructokinase
Systematic name: ADP:D-fructose-6-phosphate 1-phosphotransferase
Comments: ADP can be replaced by GDP, ATP and GTP, to a limited extent. Divalent cations are necessary for activity, with Mg2+ followed by Co2+ being the most effective.
References:
1. Tuininga, J.E., Verhees, C.H., van der Oost, J., Kengen, S.W., Stams, A.J. and de Vos, W.M. Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 274 (1999) 21023-21028. [PMID: 10409652]
Common name: ADP-specific glucokinase
Reaction: ADP + D-glucose = AMP + D-glucose 6-phosphate
Other name(s): ADP-dependent glucokinase
Systematic name: ADP:D-glucose 6-phosphotransferase
Comments: Requires Mg2+. The enzyme from Pyrococcus furiosus is highly specific for D-glucose; there is some activity with 2-deoxy-D-glucose, but no activity with D-fructose, D-mannose or D-galactose as the substrate. No activity is detected when ADP is replaced by ATP, GDP, phosphoenolpyruvate, diphosphate or polyphosphate.
References:
1. Kengen, S.W., Tuininga, J.E., de Bok, F.A., Stams, A.J. and de Vos, W.M. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270 (1995) 30453-30457. [PMID: 8530474]
Common name: phosphatidylcholine synthase
Reaction: CDPdiacylglycerol + choline = CMP + phosphatidylcholine
Other name(s): CDPdiglyceride-choline O-phosphatidyltransferase
Systematic name: CDPdiacylglycerol:choline O-phosphatidyltransferase
Comments: Requires divalent cations, with Mn2+ being more effective than Mg2+.
References:
1. de Rudder, K.E.E., Sohlenkamp, C. and Geiger, O. Plant-exudated choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 274 (1999) 20011-20016. [PMID: 10391951]
2. Sohlenkamp, C., de Rudder, K.E.E., Röhrs, V., López-Lara, I.M. and Geiger, O. Cloning and characterization of the gene for phosphatidylcholine synthase. J. Biol. Chem. 275 (2000) 18919-18925. [PMID: 10858449]
Common name: [heparan sulfate]-glucosamine N-sulfotransferase
Reaction: 3'-phosphoadenylyl sulfate + [heparan sulfate]-glucosamine = adenosine 3',5'-bisphosphate + [heparan sulfate]-N-sulfoglucosamine
Glossary:
PAPS, 3'-phosphoadenylyl sulfate nucleotide sulfate
Other names: heparin N-sulfotransferase; 3'-phosphoadenylylsulfate:N-desulfoheparin sulfotransferase; PAPS:N-desulfoheparin sulfotransferase; PAPS:DSH sulfotransferase; N-HSST; N-heparan sulfate sulfotransferase; heparan sulfate N-deacetylase/N-sulfotransferase; heparan sulfate 2-N-sulfotransferase; heparan sulfate N-sulfotransferase; heparan sulfate sulfotransferase; N-desulfoheparin sulfotransferase; desulfoheparin sulfotransferase; 3'-phosphoadenylyl-sulfate:N-desulfoheparin N-sulfotransferase; heparitin sulfotransferase; 3'-phosphoadenylyl-sulfate:heparitin N-sulfotransferase
Systematic name: 3'-phosphoadenylyl-sulfate:[heparan sulfate]-glucosamine N-sulfotransferase
Comments: The enzyme also catalyses the sulfation of chondroitin 4-sulfate and dermatan sulfate, but to a much more limited extent.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 9026-75-9
References:
1. Suzuki, S., Trenn, R.H. and Strominger, J.L. Separation of specific mucopolysaccharide sulfotransferases. Biochim. Biophys. Acta 50 (1961) 169-174.
2. Eisenman, R.A., Balasubramanian, A.S. and Marx, W. 3'-Phosphoadenylylsulfate:N-desulfoheparin sulfotransferase associated with a postmicrosomal particulate mastocytoma fraction. Arch. Biochem. Biophys. 119 (1967) 387-397. [PMID: 4964017]
3. Johnson, A.H. and Baker, J.R. The enzymatic sulphation of heparan sulphate by hen's uterus. Biochim. Biophys. Acta 320 (1973) 341-351. [PMID: 4270798]
[EC 2.8.2.12 Deleted entry: heparitin sulfotransferase. Enzyme identical to EC 2.8.2.8, [heparan sulfate]-glucosamine N-sulfotransferase (EC 2.8.2.12 created 1972, deleted 2001)]
Common name: [heparan sulfate]-glucosamine 3-sulfotransferase 1
Reaction: 3'-phosphoadenylyl sulfate + [heparan sulfate]-glucosamine = adenosine 3',5'-bisphosphate + [heparan sulfate]-glucosamine 3-sulfate
Glossary:
PAPS, 3'-phosphoadenylyl sulfate nucleotide sulfate
heparan sulfate: for definition click here
Other name(s): heparin-glucosamine 3-O-sulfotransferase; 3'-phosphoadenylyl-sulfate:heparin-glucosamine 3-O-sulfotransferase; glucosaminyl 3-O-sulfotransferase; heparan sulfate D-glucosaminyl 3-O-sulfotransferase; isoform/isozyme 1 (3-OST-1, HS3ST1)
Systematic name: 3'-phosphoadenylyl-sulfate:[heparan sulfate]-glucosamine 3-sulfotransferase
Comments: This enzyme differs from the other [heparan sulfate]-glucosamine 3-sulfotransferases [EC 2.8.2.29 ([heparan sulfate]-glucosamine 3-sulfotransferase 2) and EC 2.8.2.30 ([heparan sulfate]-glucosamine 3-sulfotransferase 3)] by being the most selective for a precursor of the antithrombin-binding site. It has a minimal acceptor sequence of: GlcNAc6S
GlcA
GlcN2S*±6S
IdoA2S
GlcN2S
, the asterisk marking the target (symbols as in 2-Carb-38 using ± to mean the presence or absence of a substituent, and > to separate a predominant structure from a minor one. Thus Glc(N2S>NAc) means a residue of glucosamine where the N carries a sulfo group mainly but occasionally an acetyl group.) [1-4]. It can also modify other precursor sequences within heparan sulfate but this action does not create functional antithrombin-binding sites. These precursors are variants of the consensus sequence:
Glc(N2S>NAc)±6S
GlcA
GlcN2S*±6S
GlcA>IdoA±2S
Glc(N2S/NAc)±6S
[5]. If the heparan sulfate substrate lacks 2-O-sulfation of GlcA residues, then enzyme specificity is expanded to modify selected glucosamine residues preceded by IdoA as well as GlcA [6].
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 118113-79-4
References:
1. Kusche, M., Backström, G., Riesenfeld, J., Pepitou, M., Choay, J. and Lindahl, U. Biosynthesis of heparin. O-Sulfation of the antithrombin-binding region. J. Biol. Chem. 263 (1988) 15474-15484. [PMID: 3139669]
2. Shworak, N.W., Fritze, L.M.S., Liu, J., Butler, L.D. and Rosenberg, R.D. Cell-free synthesis of anticoagulant heparan sulfate reveals a limiting activity which modifies a nonlimiting precursor pool. J. Biol. Chem. 271 (1996) 27063-27071. [PMID: 8900197]
3. Liu, J., Shworak, N.W., Fritze, L.M.S., Edelberg, J.M. and Rosenberg, R.D. Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J. Biol. Chem. 271 (1996) 27072-27082. [PMID: 8900198]
4. Shworak, N.W., Liu, J., Fritze, L.M.S., Schwartz, J.J., Zhang, L., Logeart, D. and Rosenberg, R.D. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J. Biol. Chem. 272 (1997) 28008-28019. [PMID: 9346953]
5. Zhang, L., Yoshida, K., Liu, J. and Rosenberg, R.D. Anticoagulant heparan sulfate precursor structures in F9 embryonal carcinoma cells. J. Biol. Chem. 274 (1999) 5681-5691 [PMID: 10026187]
6. Zhang, L., Lawrence, R., Schwartz, J.J., Bai, X., Wei., G, Esko, J.D. and Rosenberg, R.D. The effect of precursor structures on the action of glucosaminyl 3-O-sulfotransferase-1 and the biosynthesis of anticoagulant heparan sulfate. J. Biol. Chem. 276 (2001) 28806-28813. [PMID: 11375390]
Common name: [heparan sulfate]-glucosamine 3-sulfotransferase 2
Reaction: 3'-phosphoadenylyl sulfate + [heparan sulfate]-glucosamine = adenosine 3',5'-bisphosphate + [heparan sulfate]-glucosamine 3-sulfate
Glossary:
PAPS, 3'-phosphoadenylyl sulfate nucleotide sulfate
heparan sulfate: for definition click here
Other name(s): glucosaminyl 3-O-sulfotransferase; heparan sulfate D-glucosaminyl 3-O-sulfotransferase; isoform/isozyme 2 (3-OST-2, HS3ST2)
Systematic name: 3'-phosphoadenylyl-sulfate:[heparan sulfate]-glucosamine 3-sulfotransferase
Comments: This enzyme sulfates the residues marked with an asterisk in sequences containing at least IdoA2S
GlcN*
or
GlcA2S
GlcN*
(symbols as in 2-Carb-38). Preference for GlcN2S vs. unmodified GlcN has not yet been established. Additional structural features are presumably required for substrate recognition, since the 3-O-sulfated residue is of low abundance, whereas the above IdoA-containing sequence is quite abundant. This enzyme differs from the other [heparan sulfate]-glucosamine 3-sulfotransferases by modifying selected glucosamine residues preceded by GlcA2S; EC 2.8.2.23 ([heparan sulfate]-glucosamine 3-sulfotransferase 1) prefers GlcA or IdoA, whereas EC 2.8.2.30 ([heparan sulfate]-glucosamine 3-sulfotransferase 3) prefers IdoA2S.
References:
1. Shworak, N.W., Liu, J., Petros, L.M., Copeland, N.G., Jenkins N.A. and Rosenberg, R.D. Diversity of the extensive heparan sulfate D-glucosaminyl 3-O-sulfotransferase (3-OST) multigene family. J. Biol. Chem. 274 (1999) 5170-5184. [PMID: 9988767]
2. Liu, J., Shworak, N.W., Sina, P., Schwartz, J.J., Zhang, L., Fritze, L.M.S. and Rosenberg, R.D. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J. Biol. Chem. 274 (1999) 5185-5192. [PMID: 9988768]
Common name: [heparan sulfate]-glucosamine 3-sulfotransferase 3
Reaction: 3'-phosphoadenylyl sulfate + [heparan sulfate]-glucosamine = adenosine 3',5'-bisphosphate + [heparan sulfate]-glucosamine 3-sulfate
Glossary:
PAPS, 3'-phosphoadenylyl sulfate nucleotide sulfate
heparan sulfate: for definition click here
Other name(s): glucosaminyl 3-O-sulfotransferase 3a, 3b; heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A; isoform/isozyme 3a and 3b (3-OST-3A, 3-OST-3B, HS3ST3A, HS3ST3B)
Systematic name: 3'-phosphoadenylyl-sulfate:[heparan sulfate]-glucosamine 3-sulfotransferase
Comments: Two major substrates contain the tetrasaccharides: undetermined 2-sulfo-uronic acid
GlcN2S
IdoA2S
GlcN*
and
undetermined 2-sulfo-uronic acid
GlcN2S
IdoA2S
GlcN6S*
(symbols as in 2-Carb-38) with modification of the N-unsubstituted glucosamine residue (shown with an asterisk) [1,4]. Modification of selected sequences containing N-sulfo-glucosamine residues cannot yet be excluded. The 3-O-sulfated heparan sulfate can be utilized by Herpes simplex virus type 1 as an entry receptor to infect the target cells [2]. There are two isozymes, known as 3-OST-3A and 3-OST-3B, which have identical catalytic domains but are encoded by different mammalian genes [3]. The specificity of this enzyme differs from that of the other [heparan sulfate]-glucosamine 3-sulfotransferases. It is inefficient at modifying precursors of the antithrombin binding site [in contrast to EC 2.8.2.23 ([heparan sulfate]-glucosamine 3-sulfotransferase 1)] and it does not modify glucosamine preceded by GlcA2S [unlike EC 2.8.2.29 ([heparan sulfate]-glucosamine 3-sulfotransferase 2)].
References:
1. Liu, J., Shriver, Z., Blaiklock, P., Yoshida, K., Sasisekharan, R. and Rosenberg, R.D. Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A sulfates N-unsubstituted glucosamine. J. Biol. Chem. 274 (1999) 38155-38162. [PMID: 10608887]
2. Shukla, D., Liu, J., Blaiklock, P., Shworak, N.W., Bai, X., Esko, J.D., Cohen, G.H., Eisenberg, R.J., Rosenberg, R.D. and Spear, P.G. A novel role for 3-O-sulfated heparan sulfate in Herpes simplex virus 1 entry. Cell 99 (1999) 13-22. [PMID: 10520990]
3. Shworak, N.W., Liu, J., Petros, L.M., Copeland, N.G., Jenkins N.A. and Rosenberg, R.D. Diversity of the extensive heparan sulfate D-glucosaminyl 3-O-sulfotransferase (3-OST) multigene family. J. Biol. Chem. 274 (1999) 5170-5184. [PMID: 9988767]
4. Liu, J., Shworak, N.W., Sina, P., Schwartz, J.J., Zhang, L., Fritze, L.M.S. and Rosenberg, R.D. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J. Biol. Chem. 274 (1999) 5185-5192. [PMID: 9988768]
EC 2.8.4 Transferring alkylthio groups
Common name: coenzyme-B sulfoethylthiotransferase
Reaction: 2-(methylthio)ethanesulfonate (methyl-CoM) + N-(7-mercaptoheptanoyl)threonine 3-O-phosphate (coenzyme B) = CoM-S-S-CoB + methane
For diagram click here.
Glossary:
coenzyme B (CoB) is N-(7-mercaptoheptanoyl)threonine 3-O-phosphate
coenzyme M (CoM) is 2-mercaptoethanesulfonate
Other name(s): methyl-CoM reductase; methyl coenzyme M reductase
Systematic name: 2-(methylthio)ethanesulfonate:N-(7-thioheptanoyl)-3-O-phosphothreonine S-(2-sulfoethyl)thiotransferase
Comments: The enzyme from methanogenic bacteria requires the hydroporphinoid nickel complex coenzyme F430. Highly specific for coenzyme B with a heptanoyl chain; ethyl CoM and difluoromethyl CoM are poor substrates. The sulfide sulfur can be replaced by selenium but not by oxygen.
References:
1. Bobik, T.A., Olson, K.D., Noll, K.M. and Wolfe, R.S. Evidence that the heterodisulfide of coenzyme-M and 7-mercaptanoylthreonine phosphate is a product of the methylreductase reaction in Methanobacterium. Biochem. Biophys. Res. Commun. 149 (1987) 455-460. [PMID: 3122735]
2. Ellermann, J., Hedderich, R., Boecher, R. and Thauer, R.K. The final step in methane formation: investigations with highly purified methyl coenzyme M reductase component C from Methanobacterium thermoautotrophicum (strain Marburg). Eur. J. Biochem. 184 (1988) 63-68.
3. Ermler, U., Grabarse, W., Shima, S., Goubeaud, M. and Thauer, R.K. Crystal structure of methyl coenzyme M reductase: The key enzyme of biological methane formation. Science 278 (1997) 1457-1462. [PMID: 9367957]
4. Signor, L., Knuppe, C., Hug, R., Schweizer, B., Pfaltz, A. and Jaun, B. Methane formation by reaction of a methyl thioether with a photo-excited nickel thiolate - a process mimicking methanogenesis in Archaea. Chem. Eur. J. 6 (2000) 3508-3516. [PMID: 11072815]
Common name: poly(3-hydroxybutyrate) depolymerase
Reaction: poly[(R)-3-hydroxybutanoate]n + H2O = poly[(R)-3-hydroxybutanoate]n-x + poly[(R)-3-hydroxybutanoate]
Other name(s): PHB depolymerase, poly(3HB) depolymerase; poly[(R)-hydroxyalkanoic acid] depolymerase; poly(HA) depolymerase; poly(HASCL) depolymerase
Systematic name: poly[(R)-3-hydroxybutyrate] hydrolase
Comments: Reaction also occurs with esters of other short-chain-length (C1-C5) hydroxyalkanoic acids (HA). There are two types of polymers: native (intracellular) granules are amorphous and have an intact surface layer; denatured (extracellular) granules either have no surface layer or a damaged surface layer and are partially crystalline.
References:
1. Jendrossek, D. Microbial degradation of polyesters. Adv. Biochem. Eng./Biotechnol. 71 (2001) 293-325. [PMID: 11217416]
2. García, B., Olivera, E.R., Miñambres, B., Fernández-Valverde, Cañedo, L.M., Prieto, M.A., García, J.L., Martínez, M. and Luengo, J.M. Novel biodegradable aromatic plastics from a bacterial source. Genetic and biochemical studies on a route of the phenylacetyl-CoA catabolon. J. Biol. Chem. 274 (1999) 29228-29241. [PMID: 10506180]
Common name: poly(3-hydroxyoctanoate) depolymerase
Reaction: poly[(R)-3-hydroxyoctanoate]n + H2O = poly[(R)-3-hydroxyoctanoate]n-x + poly[(R)-3-hydroxyoctanoate]x; x = 1-5
Other name(s): PHO depolymerase, poly(3HO) depolymerase; poly[(R)-hydroxyalkanoic acid] depolymerase; poly(HA) depolymerase; poly(HAMCL) depolymerase
Systematic name: poly[(R)-3-hydroxyoctanoate] hydrolase
Comments: Reaction also occurs with other medium-chain-length (C6-C12) hydroxyalkanoic acids. The enzyme can also hydrolyse copolymers of medium-chain-length (MCL) hydroxyalkanoic acids and p-nitrophenyl esters of fatty acids, but short-chain-length hydroxyalkenoic acids such as poly[(R)-3-hydroxybutanoic acid] and poly[(R)-3-hydroxypentanoic acid] are not hydrolysed. There are two types of polymers: native (intracellular) granules are amorphous and have an intact surface layer; denatured (extracellular) granules either have no surface layer or a damaged surface layer and are partially crystalline.
References:
1. Jendrossek, D. Microbial degradation of polyesters. Adv. Biochem. Eng./Biotechnol. 71 (2001) 293-325. [PMID: 11217416]
2. García, B., Olivera, E.R., Miñambres, B., Fernández-Valverde, Cañedo, L.M., Prieto, M.A., García, J.L., Martínez, M. and Luengo, J.M. Novel biodegradable aromatic plastics from a bacterial source. Genetic and biochemical studies on a route of the phenylacetyl-CoA catabolon. J. Biol. Chem. 274 (1999) 29228-29241. [PMID: 10506180]
3. Schirmer, A., Jendrossek, D. and Schlegel, H.G. Degradation of poly(3-hydroxyoctanoic acid) [P(3HO)] by bacteria: putification and properties of a P(3HO) depolymerase from Pseudomonas fluorescens GK13. Appl. Environ. Microbiol. 59 (1993) 1220-1227. [PMID: 8476295]
Common name: acyloxyacyl hydrolase
Reaction: 3-(acyloxy)acyl group of bacterial toxin = 3-hydroxyacyl group of bacterial toxin + a fatty acid
For diagram click here.
Comments: The substrate is lipid A on the reducing end of the toxic lipopolysaccharide (LPS) of Salmonella typhimurium and related organisms. It consists of diglucosamine, β-D-GlcN-(1 6)-D-GlcN, attached by glycosylation on O-6 of its non-reducing residue, phosphorylated on O-4 of this residue and on O-1 of its potentially reducing residue. Both residues carry 3-(acyloxy)acyl groups on N-2 and O-3. The enzyme from human leucocytes detoxifies the lipid by hydrolysing the secondary acyl groups from O-3 of the 3-hydroxyacyl groups on the disaccharide (LPS). It also possesses a wide range of phospholipase and acyltransferase activities [e.g. EC 3.1.1.4 (phospholipase A2), EC 3.1.1.5 (lysophospholipase), EC 3.1.1.32 (phospholipase A1) and EC 3.1.1.52 (phosphatidylinositol deacylase)], hydrolysing diacylglycerol and phosphatidyl compounds, but not triacylglycerols. It has a preference for saturated C12-C16 acyl groups.
References:
1. Erwin, A.L. and Munford, R.S. Deacylation of structurally diverse lipopolysaccharides by human acyloxyacyl hydrolase. J. Biol. Chem. 265 (1990) 16444-16449. [PMID: 2398058]
2. Hagen, F.S., Grant, F.J., Kuijper, J.L., Slaughter, C.A., Moomaw, C.R., Orth, K., O'Hara, P.J. and Munford, R.S. Expression and characterization of recombinant human acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides. Biochemistry 30 (1991) 8415-8423 [PMID: 1883828]
3. Munford, R.S. and Hunter, J.P. Acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides, has phospholipase, lysophospholipase, diacylglycerollipase, and acyltransferase activities in vitro. J. Biol. Chem. 267 (1992) 10116-10121. [PMID: 1577781]
Common name: glucosylglycerol 3-phosphatase
Reaction: 2-(β-D-glucosyl)-sn-glycerol-3-phosphate + H2O = 2-(β-D-glucosyl)-sn-glycerol + phosphate
Other names: salt tolerance protein A, StpA
Systematic name: 2-(β-D-glucosyl)-sn-glycerol-3-phosphate phosphohydrolase
Comments: Acts with EC 2.4.1.213 (glucosylglycerol-phosphate synthase) to form glucosylglycerol, an osmolyte that endows cyanobacteria with resistance to salt.
References:
1. Hagemann, M. and Erdmann, N. Activation and pathway of glucosylglycerol biosynthesis in the cyanobacterium Synechosytis sp. PCC 6803. Microbiology 140 (1994) 1427-1431.
2. Hagemann, M., Richter, S., Zuther, E. and Schoor, A. Characterization of a glucosylglycerol-phosphate-accumulating salt-sensitive mutant of the cyanobacterium Synechocystis sp. strain PCC 6803. Arch. Microbiol. 166 (1996) 83-91. [PMID: 8772170]
3. Hagemann, M., Schoor, A., Jeanjean, R., Zuther, E. and Joset, F. The gene stpA from Synechocystis sp. strain PCC 6803 encodes for the glucosylglycerol-phosphate phosphatase involved in cyanobacterial salt adaptation. J. Bacteriol. 179 (1997) 1727-1733. [PMID: 9045835]
Common name: deoxyribonuclease V
Reaction: Endonucleolytic cleavage at apurinic or apyrimidinic sites to products with a 5'-phosphate
Other name(s): endodeoxyribonuclease V; DNase V; Escherichia coli endodeoxyribonuclease V
Comments: Previously classified erroneously as EC 3.1.22.3.
References:
1. Gates, F.T. and Linn, S. Endonuclease V of Escherichia coli. J. Biol. Chem. 252 (1977) 1647-1653. [PMID: 1415914159]
[EC 3.1.22.3 Transferred entry: now EC 3.1.21.7, deoxyribonuclease V (EC 3.1.22.3 created 1978, deleted 2001)]
Common name: cellulase
Reaction: Endohydrolysis of 1,4-β-D-glucosidic linkages in cellulose, lichenin and cereal β-D-glucans
Other name(s): endo-1,4-β-D-glucanase; β-1,4-glucanase; β-1,4-endoglucan hydrolase; celluase A; cellulosin AP; endoglucanase D; alkali cellulase; cellulase A 3; celludextrinase; 9.5 cellulase; avicelase; pancellase SS
Systematic name: 1,4-(1,3;1,4)-β-D-glucan 4-glucanohydrolase
Comments: Will also hydrolyse 1,4-linkages in β-D-glucans also containing 1,3-linkages.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 9012-54-8
References:
1. Datta, P.K., Hanson, K.R. and Whitaker, D.R. Improved procedures for preparation and characterization of Myrothecum cellulase. III. Molecular weight, amino acid composition, terminal residues, and other properties. Can. J. Biochem. Physiol. 41 (1963) 697-705.
2. Larner, J. Other glucosidases, in Boyer, P.D., Lardy, H. and Myrbäck, K. (Eds.), The Enzymes, 2nd edn., vol. 4, Academic Press, New York, 1960, pp. 369-378.
3. Myers, F.L. and Northcote, D.H. Partial purification and some properties of a cellulase from Helix pomatia. Biochem. J. 71 (1959) 749-756.
4. Nishizawa, K. and Hashimoto, Y. Cellulose splitting enzymes. VI. Difference in the specificities of cellulase and β-glucosidase from Irpex lacteus. Arch. Biochem. Biophys. 81 (1959) 211-222.
5. Whitaker, D.R., Hanson, K.R. and Datta, P.K. Improved procedures for preparation and characterization of myrothecium cellulase. 2. Purification procedures. Can. J. Biochem. Physiol. 41 (1963) 671-696.
6. Hatfield, R. and Nevins, D.J. Purification and properties of an endoglucanase isolated from the cell walls of Zea mays seedlings. Carbohydr. Res. 148 (1986) 265-278.
7. Hatfield, R. and Nevins, D.J. Hydrolytic activity and substrate specificity of an endoglucanase from Zea mays seedling cell walls. Plant Physiol. 83 (1987) 203-207.
8. Inohue, M., Hayashgi, K. and Nevins, D.J. Polypeptide characteristics and immunological properties of exo- and endoglucanases purified from maize coleoptile cell walls. J. Plant Physiol. 154 (1999) 334-340.
Common name: hyalurononglucosaminidase
Reaction: Random hydrolysis of 14-linkages between N-acetyl-β-D-glucosamine and D-glucuronate residues in hyaluronate
Other name(s): hyaluronidase; hyaluronoglucosidase; chondroitinase; chondroitinase I
Systematic name: hyaluronate 4-glycanohydrolase
Comments: Also hydrolyses 1,4-β-D-glycosidic linkages between N-acetyl-galactosamine or N-acetylgalactosamine sulfate and glucuronic acid in chondroitin, chondroitin 4- and 6-sulfates, and dermatan.
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 37326-33-3
References:
1. Meyer, K., Hoffman, P. and Linker, A. Hyaluronidases, in Boyer, P.D., Lardy, H. and Myrbäck, K. (Eds.), The Enzymes, 2nd edn., vol. 4, Academic Press, New York, 1960, pp. 447-460.
2. Rapport, M.M., Myer, K. and Linker, A. Analysis of the products formed on hydrolysis of hyaluronic acid by testicular hyaluronidase. J. Am. Chem. Soc. 73 (1951) 2416-2420.
3. Weissmann, B. The transglycosylative action of testicular hyaluronidase. J. Biol. Chem. 216 (1955) 783-794.
Common name: glucan 1,6-α-glucosidase
Reaction: Hydrolysis of (16)-α-D-glucosidic linkages in 1
6-α-D-glucans and derived oligosaccharides
Other name(s): exo-1,6-β-glucosidase; glucodextrinase
Systematic name: glucan α-1,6-D-glucohydrolase
Comments: Hydrolysis is accompanied by inversion at C-1, so that new reducing ends are released in the β-configuration.. Dextrans and isomaltosaccharides are hydrolysed, as is isomaltose, but very slowly. The enzyme from some sources also possesses the activity of EC 3.2.1.59 (glucan endo-1,3-α-glucosidase).
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 37288-48-5
References:
1. Ohya, T., Sawai, T., Uemura, S. and Abe, K. Some catalytic properties of an exo-1,6-α-glucosidase (glucodextranase) from Arthrobacter globiformis I42. Agric. Biol. Chem. 42 (1978) 571-577.
2. Sawai, T., Yamaki, T. and Ohya, T. Preparation and some properties of Arthrobacter globiformis exo-1,6-α-glucosidase. Agric. Biol. Chem. 40 (1976) 1293-1299.
3. Walker, G.J. and Pulkownik, A. Degradation of dextrans by an α-1,6-glucan glucohydrolase from Streptococcus mitis. Carbohydr. Res. 29 (1973) 1-14. [PMID: 4356399]
Common name: β-primeverosidase
Reaction: a 6-O-(β-D-xylopyranosyl)-β-D-glucopyranoside + H2O = 6-O-(β-D-xylopyranosyl)-β-D-glucopyranose + an alcohol
Glossary
primeverose = 6-O-(β-D-xylopyranosyl)-D-glucose
vicianose = 6-O-(α-L-arabinopyranosyl)-D-glucose
Systematic name: 6-O-(β-D-xylopyranosyl)-β-D-glucopyranoside 6-O-(β-D-xylosyl)-β-D-glucohydrolase
Comments: The enzyme is responsible for the formation of the alcoholic aroma in oolong and black tea. In addition to β-primeverosides [i.e. 6-O-(β-D-xylopyranosyl)-β-D-glucopyranosides], it also hydrolyses 6-O-(β-D-apiofuranosyl)-β-D-glucopyranosides and, less rapidly, β-vicianosides and 6-O-(α-L-arabinofuranosyl)-β-D-glucopyranosides, but not β-glucosides. Geranyl-, linaloyl-, benzyl- and p-nitrophenol glycosides are all hydrolysed.
References
1. Ijima, Y., Ogawa, K., Watanabe, N., Usui, T., Ohnishi-Kameyama, M., Nagata, T. and Sakata, K. Characterization of β-primeverosidase, being concerned with alcoholic aroma formation in tea leaves to be processed into black tea, and preliminary observations on its substrate specificity. J. Agric. Food Chem. 46 (1998) 1712-1718.
2. Ogawa, K., Ijima, Y., Guo, W., Watanabe, N., Usui, T., Dong, S., Tong, Q. and Sakata, K. Purification of a β-primeverosidase concerned with alcoholic aroma formation in tea leaves (cv. Shuxian) to be processed to oolong tea. J. Agric. Food Chem. 45 (1997) 877-882.
Common name: N-carbamoyl-L-amino-acid hydrolase
Reaction: N-carbamoyl-L-2-amino acid (a 2-ureido carboxylate) + H2O = L-2-amino acid + NH3 + CO2
Systematic name: N-carbamoyl-L-amino acid amidohydrolase
Comments: The enzyme has broad specificity for carbamoyl-L-amino acids, although it is inactive on the carbamoyl derivatives of glutamate, aspartate, arginine, tyrosine or tryptophan. Carbamoyl-L-valine is the best substrate. It is inactive on derivatives of D-amino acids, the substrates of EC 3.5.1.77 (N-carbamoyl-D-amino acid hydrolase), or on N-carbamoyl-β-alanine, the substrate of EC 3.5.1.6 (β-ureidopropionase). The enzyme hydrolyses formyl derivatives rapidly, and acetyl derivatives very slowly. Mn2+, Ni2+ or Co2+ enabled reactivation of enzyme that had been treated with a chelating agent.
References:
1. Ogawa, J., Miyake, H. and Shimizu, S. Purification and characterization of N-carbamoyl-L-amino acid amidohydrolase with broad substrate specificity from Alcaligenes xylosoxidans. Appl. Microbiol. Biotechnol. 43 (1995) 1039-1043. [PMID: 8590654]
EC 3.13 Acting on Carbon-Sulfur Bonds
Common name: UDPsulfoquinovose synthase
Reaction: UDPglucose + sulfite = UDP-6-sulfoquinovose + H2O
For diagram click here.
Other name(s): sulfite:UDP-glucose sulfotransferase
Systematic name: UDP-6-sulfo-6-deoxyglucose sulfohydrolase
Comments: Requires NAD+, which appears to oxidize the substrate to UDP-4-dehydroglucose, which dehydrates to UDP-4-dehydro-6-deoxygluc-5-enose, to which sulfite can add; the reaction is completed when the substrate is rehydrogenated at C-4. The enzyme from Arabidopsis thaliana is specific for UDP-Glc and sulfite.
References:
1. Essigmann, B., Gler, S., Narang, R.A., Linke, D. and Benning, C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 (1998) 1950-1955. [PMID: 9465123]
2. Essigmann, B., Hespenheide, B.M., Kuhn, L.A. and Benning, C. Prediction of the active-site structure and NAD+ binding in SQD1, a protein essential for sulfolipid biosynthesis in Arabidopsis. Arch. Biochem. Biophys. 369 (1999) 30-41. [PMID: 10462438]
3. Mulichak, A.M., Theisen, M.J., Essigmann, B., Benning, C. and Garavito, R.M. Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose. Proc. Natl. Acad. Sci. USA 96 (1999) 13097-13102. [PMID: 10557279]
4. Sanda, S., Leustek, T., Theisen, M., Garavito, R.M. and Benning, C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 276 (2001) 3941-3946. [PMID: 11073956]
Common name: 1-deoxy-D-xylulose 5-phosphate synthase
Reaction: pyruvate + D-glyceraldehyde 3-phosphate = 1-deoxy-D-xylulose 5-phosphate + CO2
For diagram click here.
Other name(s): DXP-synthase
Systematic name: 1-deoxy-D-xylulose-5-phosphate pyruvate-lyase (carboxylating)
Comments: Requires thiamine diphosphate. The enzyme catalyses a step in the nonmevalonate pathway of terpenoid synthesis; the next step is catalysed by EC 1.1.1.267 (1-deoxy-D-xylulose-5-phosphate reductoisomerase).
References:
1. Kuzuyama, T., Takagi, M., Takahashi, S. and Seto, H. Cloning and characterization of 1-deoxy-D-xylulose 5-phosphate synthase from Streptomyces sp. strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J. Bacteriol. 182 (2000) 891-897. [PMID: 10648511]
[EC 4.2.1.13 Transferred entry: now EC 4.3.1.17, L-serine ammonia-lyase (EC 4.2.1.13 created 1961, deleted 2001)]
[EC 4.2.1.14 Transferred entry: now EC 4.3.1.18, D-serine ammonia-lyase (EC 4.2.1.14 created 1961, deleted 2001)]
[EC 4.2.1.16 Transferred entry: now EC 4.3.1.19, threonine ammonia-lyase (EC 4.2.1.16 created 1961, deleted 2001)]
[EC 4.2.1.38 Transferred entry: now EC 4.3.1.20, erythro-3-hydroxyaspartate ammonia-lyase (EC 4.2.1.38 created 1972, deleted 2001)]
Common name: cyclohexyl-isocyanide hydratase
Reaction: N-cyclohexylformamide = cyclohexyl isocyanide + H2O
Other name(s): isonitrile hydratase
Systematic name: N-cyclohexylformamide hydro-lyase
Comments: The enzyme from Pseudomonas putida strain N19-2 can also catalyse the hydration of other isonitriles to the corresponding N-substituted formamides. The enzyme has no metal requirements.
References:
1. Goda, M., Hashimoto, Y., Shimizu, S. and Kobayashi, M. Discovery of a novel enzyme, isonitrile hydratase, involved in nitrogen-carbon triple bond cleavage. J. Biol. Chem. 276 (2001) 23480-23485. [PMID: 11306561]
Common name: hyaluronate lyase
Reaction: hyaluronate = n 3-(4-deoxy-β-D-gluc-4-enuronosyl)-N-acetyl-D-glucosamine
Other name(s): hyaluronidase [but cf. EC 3.2.1.35 (hyalurononglucosaminidase) and EC 3.2.1.36 (hyaluronoglucuronidase)]; glucuronoglycosaminoglycan lyase; spreading factor; mucinase
Systematic name: hyaluronate lyase
Comments: Also acts on chondroitin. Formerly EC 4.2.99.1. The product is more systematically known as 3-(4-deoxy-α-L-threo-hex-4-enopyranosyluronic acid)-2-acetamido-2-deoxy-D-glucose
Links to other databases: BRENDA, EXPASY, KEGG, WIT, CAS registry number: 37259-53-3
References:
1. Linker, A., Hoffman, P., Meyer, K., Sampson, P. and Korn, E.D. The formation of unsaturated disacharides from mucopoly-saccharides and their cleavage to α-keto acid by bacterial enzymes. J. Biol. Chem. 235 (1960) 3061.
2. Meyer, K. and Rapport, M.M. Hyaluronidases. Adv. Enzymol. Relat. Subj. Biochem. 13 (1952) 199-236.
3. Moran, F., Nasuno, S. and Starr, M.P. Extracellular and intracellular polygllacturonic acid trans-eliminases of Erwinia carotovora. Arch. Biochem. Biophys. 123 (1968) 298-306. [Medline UI: 68165141]
*EC 4.3 Carbon-Nitrogen Lyases
This subclass contains the enzymes that release ammonia or one of its derivatives, with the formation of a double bond or ring. Some catalyse the actual elimination of the ammonia, amine or amide, e.g.
Others, however, catalyse elimination of another component, e.g. water, which is followed by spontaneous reactions that lead to breakage of the C-N bond, e.g.
as in EC 4.3.1.17 (L-serine ammonia-lyase), so that the overall reaction is
i.e., an elimination with rearrangement. The sub-subclasses of EC 4.3 are the ammonia-lyases (EC 4.3.1), the amidine-lyases (EC 4.3.2), the amine-lyases (EC 4.3.3), and other carbon-nitrogen lyases (EC 4.3.99).
Common name: threo-3-hydroxyaspartate ammonia-lyase
Reaction: threo-3-hydroxy-L-aspartate = oxaloacetate + NH3
Other name(s): threo-3-hydroxyaspartate dehydratase; L-threo-3-hydroxyaspartate dehydratase
Systematic name: threo-3-hydroxy-L-aspartate ammonia-lyase
Comments: A pyridoxal-phosphate protein.
References:
1. Wada, M., Matsumoto, T., Nakamori, S., Sakamoto, M., Kataoka, M., Liu, J.-Q., Itoh, N., Yamada, H. and Shimizu, S. Purification and characterization of a novel enzyme, L-threo-3-hydroxyaspartate dehydratase, from Pseudomonas sp. T62. FEMS Microbiol. Lett. 179 (1999) 147-151. [PMID: 10481099]
Common name: L-serine ammonia-lyase
Reaction: L-serine = pyruvate + NH3
Other name(s): serine deaminase; L-hydroxyaminoacid dehydratase; L-serine deaminase; L-serine dehydratase; L-serine hydro-lyase (deaminating)
Systematic name: L-serine ammonia-lyase
Comments: A pyridoxal-phosphate protein. This reaction is also carried out by EC 4.3.1.19 threonine ammonia-lyase, from a number of sources. The reaction catalysed probably involves initial elimination of water (hence the enzyme's original classification as EC 4.2.1.13, L-serine dehydratase), followed by isomerization and hydrolysis of the product with C-N bond breakage.
References:
1. Ramos, F. and Wiame, J.-M. Occurrence of a catabolic L-serine (L-threonine) deaminase in Saccharomyces cerevisiae. Eur. J. Biochem. 123 (1982) 571-576. [PMID: 7042346]
2. Simon, D., Hoshino, J. and Kröger, H. L-Serine dehydratase from rat liver. Purification and some properties. Biochim. Biophys. Acta 321 (1973) 361-368. [PMID: 4750769]
3. Suda, M. and Nakagawa, H. L-Serine dehydratase (rat liver). Methods Enzymol. 17B (1971) 346-351.
4. Sagers, R.D. and Carter, J. E. L-Serine dehydratase (Clostridium acidiurica). Methods Enzymol. 17B (1971) 351-356.
5. Robinson, W.G. and Labow, R. L-Serine dehydratase (Escherichia coli). Methods Enzymol. 17B (1971) 356-360.
Common name: D-serine ammonia-lyase
Reaction: D-serine = pyruvate + NH3
Other name(s): D-hydroxyaminoacid dehydratase; D-serine dehydrase; D-hydroxy amino acid dehydratase; D-serine hydrolase; D-serine dehydratase (deaminating); D-serine deaminase; D-serine hydro-lyase (deaminating)
Systematic name: D-serine ammonia-lyase
Comments: A pyridoxal-phosphate protein. Also acts, slowly, on D-threonine. The reaction catalysed probably involves initial elimination of water (hence the enzyme's original classification as EC 4.2.1.14, D-serine dehydratase), followed by isomerization and hydrolysis of the product with C-N bond breakage.
References:
1. Dupourque, D., Newton, W.A. and Snell, E.E. Purification and properties of D-serine dehydrase from Escherichia coli. J. Biol. Chem. 241 (1966) 1233-1238. [PMID: 5327100]
2. Metzler, D.E. and Snell, E.E. Deamination of serine. II. D-Serine dehydrase, a vitamin B6 enzyme from Escherichia coli. J. Biol. Chem. 198 (1952) 363-373.
Common name: threonine ammonia-lyase
Reaction: L-threonine = 2-oxobutanoate + NH3
Other name(s): threonine deaminase; L-serine dehydratase; serine deaminase; L-threonine dehydratase; threonine dehydrase; L-threonine deaminase; threonine dehydratase; L-threonine hydro-lyase (deaminating)
Systematic name: L-threonine ammonia-lyase
Comments: The enzyme from many sources is a pyridoxal-phosphate protein; that from P. putida is not. The enzyme from a number of sources also acts on L-serine, cf. EC 4.3.1.17, L-serine ammonia-lyase. The reaction catalysed probably involves initial elimination of water (hence the enzyme's original classification as EC 4.2.1.16, threonine dehydratase), followed by isomerization and hydrolysis of the product with C-N bond breakage.
References:
1. Cohn, M.S. and Phillips, A.T. Purification and characterization of a B6-independent threonine dehydratase from Pseudomonas putida. Biochemistry 13 (1974) 1208-1214. [PMID: 4814721]
2. Nishimura, J.S. and Greenberg, D.M. Purification and properties of L-threonine dehydrase of sheep liver. J. Biol. Chem. 236 (1961) 2684-2691.
3. Phillips, A.T. and Wood, W.A. The mechanism of action of 5'-adenylic acid-activated threonine dehydrase. J. Biol. Chem. 240 (1965) 4703-4309. [PMID: 5321308]
4. Shizuta, Y., Nakazawa, A., Tokushige, M. and Hayaishi, O. Studies on the interaction between regulatory enzymes and effectors. 3. Crystallization and characterization of adenosine 5'-monophosphate-dependent threonine deaminase from Escherichia coli. J. Biol. Chem. 244 (1969) 1883-1889. [PMID: 4889010]
Common name: erythro-3-hydroxyaspartate ammonia-lyase
Reaction: erythro-3-hydroxy-L-aspartate = oxaloacetate + NH3
Other name(s): 3-hydroxyaspartate dehydratase; erythro-β-hydroxyaspartate dehydratase; erythro-3-hydroxyaspartate dehydratase; erythro-3-hydroxy-Ls-aspartate hydro-lyase (deaminating)
Systematic name: erythro-3-hydroxy-Ls-aspartate ammonia-lyase
Comments: A pyridoxal-phosphate protein. The reaction catalysed probably involves initial elimination of water (hence the enzyme's original classification as EC 4.2.1.38, erythro-3-hydroxyaspartate dehydratase), followed by isomerization and hydrolysis of the product with C-N bond breakage.
References:
1. Gibbs, R.G. and Morris, J.G. Purification and properties of erythro-β-hydroxyaspartate dehydratase from Micrococcus denitrificans. Biochem. J. 97 (1965) 547-554.
Common name: D-alanine-poly(phosphoribitol) ligase
Reaction: ATP + D-alanine + poly(ribitol phosphate) = AMP + diphosphate + O-D-alanyl-poly(ribitol phosphate)
Other name(s): D-alanyl-poly(phosphoribitol) synthetase; D-alanine: membrane acceptor ligase; D-alanine-D-alanyl carrier protein ligase; D-alanine-membrane acceptor ligase; D-alanine-activating enzyme
Systematic name: D-alanine:poly(phosphoribitol) ligase (AMP-forming)
Comments: A thioester bond is formed transiently between D-alanine and the sulfhydryl group of the 4'-phosphopantetheine prosthetic group of D-alanyl carrier protein during the activation of the alanine. Involved in the synthesis of teichoic acids.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
WIT,
CAS registry number: 9023-65-8
References:
1. Baddiley, J. and Neuhaus, F.C. The enzymic activation of D-alanine. Biochem. J. 75 (1960) 579.
2. Reusch, V.M. and Neuhaus, F.C. D-Alanine:membrane acceptor ligase from Lactobacillus casei. J. Biol. Chem. 246 (1971) 6136-6143. [PMID: 4399593]
3. Perego, M., Glaser, P., Minutello, A., Strauch, M.A., Leopold, K. and Fischer, W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. J. Biol. Chem. 270 (1995) 15598-15606. [PMID: 7797557]
4. Heaton, M.P. and Neuhaus, F.C. Role of D-alanyl carrier protein in the biosynthesis of D-alanyl-lipoteichoic acid. J. Bacteriol. 176 (1994) 681-690. [PMID: 8300523]
5. Debabov, D.V., Heaton, M.P., Zhang, Q., Stewart, K.D., Lambalot, R.H. and Neuhaus, F.C. The D-alanyl carrier protein in Lactobacillus casei: cloning, sequencing and expression of dltC. J. Bacteriol. 178 (1996) 3869-3876. [PMID: 8682792]
[EC 6.2.1.21 Deleted entry: phenylacetate-CoA ligase. Activity covered by EC 6.2.1.30, phenylacetate-CoA ligase (EC 6.2.1.21 created 1986, deleted 2001)]
>C=CH- + NH2-R
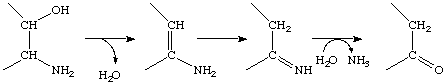
>CH2-CO- + NH3
Return to Enzymes Home Page.