Accepted name: sulfolactaldehyde 3-reductase
Reaction: 2,3-dihydroxypropane-1-sulfonate + NAD+ = 2-hydroxy-3-oxopropane-1-sulfonate + NADH + H+
For diagram of reaction click here.
Glossary: 2-hydroxy-3-oxopropane-1-sulfonate = sulfolactaldehyde
Other name(s): yihU-(gene name)
Systematic name: 2,3-dihydroxypropane-1-sulfonate:NAD+ 3-oxidoreductase
Comments: The enzyme, characterized from the bacterium Escherichia coli, is involved in the degradation pathway of sulfoquinovose, the polar headgroup of sulfolipids found in the photosynthetic membranes of all higher plants, mosses, ferns, algae, and most photosynthetic bacteria, as well as the surface layer of some archaea.
References:
1. Denger, K., Weiss, M., Felux, A.K., Schneider, A., Mayer, C., Spiteller, D., Huhn, T., Cook, A.M. and Schleheck, D. Sulphoglycolysis in Escherichia coli-K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507 (2014) 114-117. [PMID: 24463506]
[EC 1.1.3.3 Deleted entry: malate oxidase. Now classified as EC 1.1.5.4, malate dehydrogenase (quinone). (EC 1.1.3.3 created 1961, deleted 2014)]
*EC 1.1.3.19
Accepted name: 4-hydroxymandelate oxidase (decarboxylating)
Reaction: (S)-4-hydroxymandelate + O2 = 4-hydroxybenzaldehyde + CO2 + H2O2
Glossary: (S)-4-hydroxymandelate = (S)-2-hydroxy-2-(4-hydroxyphenyl)acetate
Other name(s): L-4-hydroxymandelate oxidase (decarboxylating); (S)-2-hydroxy-2-(4-hydroxyphenyl)acetate:oxygen 1-oxidoreductase; (S)-4-hydroxymandelate:oxygen 1-oxidoreductase; 4-hydroxymandelate oxidase
Systematic name: (S)-4-hydroxymandelate:oxygen 1-oxidoreductase (decarboxylating)
Comments: A flavoprotein (FAD), requires Mn2+. The enzyme from the bacterium Pseudomonas putida-is involved in the degradation of mandelate.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number: 60976-30-9
References:
1. Bhat, S.G. and Vaidyanathan, C.S. Purification and properties of L-4-hydroxymandelate oxidase from Pseudomonas convexa. Eur. J. Biochem. 68 (1976) 323-331. [PMID: 976259]
EC 1.1.3.46
Accepted name: 4-hydroxymandelate oxidase
Reaction: (S)-4-hydroxymandelate + O2 = 2-(4-hydroxyphenyl)-2-oxoacetate + H2O2
Glossary: (S)-4-hydroxymandelate = (S)-2-hydroxy-2-(4-hydroxyphenyl)acetate
Other name(s): 4HmO; HmO
Systematic name: (S)-4-hydroxymandelate:oxygen 1-oxidoreductase
Comments: A flavoprotein (FMN). The enzyme from the bacterium Amycolatopsis orientalis is involved in the biosynthesis of L-(4-hydroxyphenyl)glycine and L-(3,5-dihydroxyphenyl)glycine, two non-proteinogenic amino acids occurring in the vancomycin group of antibiotics.
References:
1. Hubbard, B.K., Thomas, M.G. and Walsh, C.T. Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem. Biol. 7 (2000) 931-942. [PMID: 11137816]
2. Li, T.L., Choroba, O.W., Charles, E.H., Sandercock, A.M., Williams, D.H. and Spencer, J.B. Characterisation of a hydroxymandelate oxidase involved in the biosynthesis of two unusual amino acids occurring in the vancomycin group of antibiotics. Chem. Commun. (Camb.) (2001) 1752-1753. [PMID: 12240298]
*EC 1.2.1.27
Accepted name: methylmalonate-semialdehyde dehydrogenase (CoA-acylating)
Reaction: 2-methyl-3-oxopropanoate + CoA + H2O + NAD+ = propanoyl-CoA + HCO3- + NADH
For diagram of reaction click here.
Glossary: methylmalonate semialdehyde = 2-methyl-3-oxopropanoate
Other name(s): MSDH; MMSA dehydrogenase; iolA (gene name); methylmalonate-semialdehyde dehydrogenase (acylating)
Systematic name: 2-methyl-3-oxopropanoate:NAD+ 3-oxidoreductase (CoA-propanoylating)
Comments: Also converts 3-oxopropanoate into acetyl-CoA [3]. The reaction occurs in two steps with the decarboxylation process preceding CoA-binding [3]. Bicarbonate rather than CO2 is released as a final product [3].
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
PDB,
CAS registry number: 37205-49-5
References:
1. Sokatch, J.R., Sanders, L.E. and Marshall, V.P. Oxidation of methylmalonate semialdehyde to propionyl coenzyme A in Pseudomonas aeruginosa grown on valine. J. Biol. Chem. 243 (1968) 2500-2506. [PMID: 4297649]
2. Dubourg, H., Stines-Chaumeil, C., Didierjean, C., Talfournier, F., Rahuel-Clermont, S., Branlant, G. and Aubry, A. Expression, purification, crystallization and preliminary X-ray diffraction data of methylmalonate-semialdehyde dehydrogenase from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 60 (2004) 1435-1437. [PMID: 15272169]
3. Stines-Chaumeil, C., Talfournier, F. and Branlant, G. Mechanistic characterization of the MSDH (methylmalonate semialdehyde
dehydrogenase) from Bacillus subtilis. Biochem. J. 395 (2006) 107-115. [PMID: 16332250]
*EC 1.3.1.10
Accepted name: enoyl-[acyl-carrier-protein] reductase (NADPH, Si-specific)
Reaction: an acyl-[acyl-carrier protein] + NADP+ = a trans-2,3-dehydroacyl-[acyl-carrier protein] + NADPH + H+
Other name(s): acyl-ACP dehydrogenase (ambiguous); enoyl-[acyl carrier protein] (reduced nicotinamide adenine dinucleotide phosphate) reductase; NADPH 2-enoyl Co A reductase; enoyl acyl-carrier-protein reductase (ambiguous); enoyl-ACP reductase (ambiguous); acyl-[acyl-carrier-protein]:NADP+ oxidoreductase (B-specific); acyl-[acyl-carrier protein]:NADP+ oxidoreductase (B-specific); enoyl-[acyl-carrier-protein] reductase (NADPH, B-specific)
Systematic name: acyl-[acyl-carrier protein]:NADP+ oxidoreductase (Si-specific)
Comments: One of the activities of EC 2.3.1.86, fatty-acyl-CoA synthase, an enzyme found in yeasts (Ascomycota and the Basidiomycota). Catalyses the reduction of enoyl-acyl-[acyl-carrier protein] derivatives of carbon chain length from 4 to 16. The yeast enzyme is Si-specific with respect to NADP+. cf. EC 1.3.1.39, enoyl-[acyl-carrier-protein] reductase (NADPH, Re-specific) and EC 1.3.1.104, enoyl-[acyl-carrier-protein] reductase (NADPH), which describes enzymes whose stereo-specificity towards NADPH is not known. See also EC 1.3.1.9, enoyl-[acyl-carrier-protein] reductase (NADH).
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
PDB,
CAS registry number: 37251-09-5
References:
1. Seyama, T., Kasama, T., Yamakawa, T., Kawaguchi, A., Saito, K. and Okuda, S. Origin of hydrogen atoms in the fatty acids synthesized with yeast fatty acid synthetase. J. Biochem. (Tokyo) 82 (1977) 1325-1329. [PMID: 338601]
EC 1.3.99.36
Accepted name: cypemycin cysteine dehydrogenase (decarboxylating)
Reaction: cypemycin(1-18)-L-Cys-L-Leu-L-Val-L-Cys + acceptor = C3.19,S21-cyclocypemycin(1-18)-L-Ala-L-Leu-N-thioethenyl-L-valinamide + CO2 + H2S + reduced acceptor
For diagram of reaction click here.
Glossary: C3.19,S21-cyclocypemycin(1-18)-L-Ala-L-Leu-N-thioethenyl-L-valinamide =
Other name(s): cypemycin decarboxylase; CypD
Systematic name: cypemycin(1-18)-L-Cys-L-Leu-L-Val-L-Cys:acceptor oxidoreductase (decarboxylating)
Comments: Cypemycin, isolated from the bacterium Streptomyces sp. OH-4156, is a peptide antibiotic, member of the linaridins, a class of posttranslationally modified ribosomally synthesized peptides. The enzyme decarboxylates and reduces the C-terminal L-cysteine residue, producing a reactive ethenethiol group that reacts with a dethiolated cysteine upstream to form an aminovinyl-methyl-cysteine loop that is important for the antibiotic activity of the mature peptide.
References:
1. Claesen, J. and Bibb, M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc. Natl. Acad. Sci. USA 107 (2010) 16297-16302. [PMID: 20805503]
EC 1.5.1.48
Accepted name: 2-methyl-1-pyrroline reductase
Reaction: (R)-2-methylpyrrolidine + NADP+ = 2-methyl-1-pyrroline + NADPH + H+
Other name(s): (R)-imine reductase (ambiguous)
Systematic name: (R)-2-methylpyrrolidine:NADP+ 2-oxidoreductase
Comments: The enzyme from the bacterium Streptomyces sp. GF3587 is highly specific for its substrate and forms only the (R) isomer.
References:
1. Mitsukura, K., Suzuki, M., Shinoda, S., Kuramoto, T., Yoshida, T. and Nagasawa, T. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci. Biotechnol. Biochem. 75 (2011) 1778-1782. [PMID: 21897027]
*EC 1.5.4.1
Accepted name: pyrimidodiazepine synthase
Reaction: 2-amino-6-acetyl-3,7,8,9-tetrahydro-3H-pyrimido[4,5-b][1,4]diazepin-4-one + glutathione disulfide + H2O = 6-pyruvoyltetrahydropterin + 2 glutathione
For diagram of reaction click here.
Glossary: 2-amino-6-acetyl-3,7,8,9-tetrahydro-3H-pyrimido[4,5-b][1,4]diazepin-4-one = pyrimidodiazepine
Other name(s): PDA synthase; pyrimidodiazepine:oxidized-glutathione oxidoreductase (ring-opening, cyclizing); pyrimidodiazepine:glutathione-disulfide oxidoreductase (ring-opening, cyclizing)
Systematic name: 2-amino-6-acetyl-3,7,8,9-tetrahydro-3H-pyrimido[4,5-b][1,4]diazepin-4-one:glutathione-disulfide oxidoreductase (ring-opening, cyclizing)
Comments: In the reverse direction, the reduction of 6-pyruvoyl-tetrahydropterin is accompanied by the opening of the 6-membered pyrazine ring and the formation of the 7-membered diazepine ring. The pyrimidodiazepine formed is an acetyldihydro derivative. Involved in the formation of the eye pigment drosopterin in Drosophila melanogaster.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number: 93586-06-2
References:
1. Wiederrecht, G.J. and Brown, G.M. Purification and properties of the enzymes from Drosophila melanogaster that catalyze the conversion of dihydroneopterin triphosphate to the pyrimidodiazepine precursor of the drosopterins. J. Biol. Chem. 259 (1984) 14121-14127. [PMID: 6438092]
2. Kim, J., Suh, H., Kim, S., Kim, K., Ahn, C. and Yim, J. Identification and characteristics of the structural gene for the Drosophila eye colour mutant sepia, encoding PDA synthase, a member of the ω class glutathione S-transferases. Biochem. J. 398 (2006) 451-460. [PMID: 16712527]
*EC 1.10.3.10
Accepted name: ubiquinol oxidase (H+-transporting)
Reaction: 2 ubiquinol + O2 + n H+[side 1] = 2 ubiquinone + 2 H2O + n H+[side 2]
Other name(s): cytochrome bb3 oxidase; cytochrome bo-oxidase; cytochrome bd-II oxidase
Systematic name: ubiquinol:O2 oxidoreductase (H+-transporting)
Comments: Contains a dinuclear centre comprising two hemes, or heme and copper. This terminal oxidase enzyme generates proton motive force by two mechanisms: (1) transmembrane charge separation resulting from utilizing protons and electrons originating from opposite sides of the membrane to generate water, and (2) active pumping of protons across the membrane. The bioenergetic efficiency (the number of charges driven across the membrane per electron used to reduce oxygen to water) depends on the enzyme; for example, for the bo3 oxidase it is 2, while for the bd-II oxidase it is 1. cf. EC 1.10.3.14, ubiquinol oxidase (electrogenic, non H+-transporting).
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number:
References:
1. Abramson, J., Riistama, S., Larsson, G., Jasaitis, A., Svensson-Ek, M., Laakkonen, L., Puustinen, A., Iwata, S. and Wikstrom, M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 7 (2000) 910-917. [PMID: 11017202]
2. Yap, L.L., Lin, M.T., Ouyang, H., Samoilova, R.I., Dikanov, S.A. and Gennis, R.B. The quinone-binding sites of the cytochrome bo3 ubiquinol oxidase from Escherichia coli. Biochim. Biophys. Acta 1797 (2010) 1924-1932. [PMID: 20416270]
3. Shepherd, M., Sanguinetti, G., Cook, G.M. and Poole, R.K. Compensations for diminished terminal oxidase activity in Escherichia coli: cytochrome bd-II-mediated respiration and glutamate metabolism. J. Biol. Chem. 285 (2010) 18464-18472. [PMID: 20392690]
4. Borisov, V.B., Murali, R., Verkhovskaya, M.L., Bloch, D.A., Han, H., Gennis, R.B. and Verkhovsky, M.I. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. USA 108 (2011) 17320-17324. [PMID: 21987791]
EC 1.10.3.15
Accepted name: grixazone synthase
Reaction: 2 3-amino-4-hydroxybenzoate + N-acetyl-L-cysteine + 2 O2 = grixazone B + 4 H2O + CO2
For diagram of reaction click here.
Glossary: grixazone B = 8-amino-9-(N-acetyl-L-cystein-S-yl)-7-oxo-7H-phenoxazine-2-carboxylic acid
Other name(s): GriF
Systematic name: 3-amino-4-hydroxybenzoate:N-acetyl-L-cysteine:oxygen oxidoreductase
Comments: A type 3 multi copper protein. The enzyme, isolated from the bacterium Streptomyces griseus, catalyses an 8 electron oxidation. Activation of the enzyme requires a copper chaperone (GriE). It also acts on 3-amino-4-hydroxybenzaldehyde, giving grixazone A. The second aldehyde group is presumably lost as formate. The enzyme also catalyses the reaction of EC 1.10.3.4 o-aminophenol oxidase.
References:
1. Suzuki, H., Ohnishi, Y., Furusho, Y., Sakuda, S. and Horinouchi, S. Novel benzene ring biosynthesis from C3 and C4 primary metabolites by two enzymes. J. Biol. Chem. 281 (2006) 36944-36951. [PMID: 17003031]
2. Le Roes-Hill, M., Goodwin, C. and Burton, S. Phenoxazinone synthase: what’s in a name. Trends Biotechnol 27 (2009) 248-258. [PMID: 19268377]
EC 1.11.1.23
Accepted name: (S)-2-hydroxypropylphosphonic acid epoxidase
Reaction: (S)-2-hydroxypropylphosphonate + H2O2 = (1R,2S)-1,2-epoxypropylphosphonate + 2 H2O
For diagram of reaction click here.
Glossary: (1R,2S)-1,2-epoxypropylphosphonate = fosfomycin = [(2R,3S)-3-methyloxiran-2-yl]phosphonate
Other name(s): HPP epoxidase; HppE; 2-hydroxypropylphosphonic acid epoxidase; Fom4; (S)-2-hydroxypropylphosphonate epoxidase
Systematic name: (S)-2-hydroxypropylphosphonate:hydrogen peroxide epoxidase
Comments: This is the last enzyme in the biosynthetic pathway of fosfomycin, a broad-spectrum antibiotic produced by certain Streptomyces species. Contains non heme iron that forms a iron(IV)-oxo (ferryl) complex with hydrogen peroxide, which functions as a proton abstractor from the substrate [7].
References:
1. Munos, J.W., Moon, S.J., Mansoorabadi, S.O., Chang, W., Hong, L., Yan, F., Liu, A. and Liu, H.W. Purification and characterization of the epoxidase catalyzing the formation of fosfomycin from Pseudomonas syringae. Biochemistry 47 (2008) 8726-8735. [PMID: 18656958]
2. Yan, F., Moon, S.J., Liu, P., Zhao, Z., Lipscomb, J.D., Liu, A. and Liu, H.W. Determination of the substrate binding mode to the active site iron of (S)-2-hydroxypropylphosphonic acid epoxidase using 17O-enriched substrates and substrate analogues. Biochemistry 46 (2007) 12628-12638. [PMID: 17927218]
3. Hidaka, T., Goda, M., Kuzuyama, T., Takei, N., Hidaka, M. and Seto, H. Cloning and nucleotide sequence of fosfomycin biosynthetic genes of Streptomyces wedmorensis. Mol. Gen. Genet. 249 (1995) 274-280. [PMID: 7500951]
4. Liu, P., Mehn, M.P., Yan, F., Zhao, Z., Que, L., Jr. and Liu, H.W. Oxygenase activity in the self-hydroxylation of (S)-2-hydroxypropylphosphonic acid epoxidase involved in fosfomycin biosynthesis. J. Am. Chem. Soc. 126 (2004) 10306-10312. [PMID: 15315444]
5. Higgins, L.J., Yan, F., Liu, P., Liu, H.W. and Drennan, C.L. Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme. Nature 437 (2005) 838-844. [PMID: 16015285]
6. Cameron, S., McLuskey, K., Chamberlayne, R., Hallyburton, I. and Hunter, W.N. Initiating a crystallographic analysis of recombinant (S)-2-hydroxypropylphosphonic acid epoxidase from Streptomyces wedmorensis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61 (2005) 534-536. [PMID: 16511089]
7. Wang, C., Chang, W.C., Guo, Y., Huang, H., Peck, S.C., Pandelia, M.E., Lin, G.M., Liu, H.W., Krebs, C. and Bollinger, J.M., Jr. Evidence that the fosfomycin-producing epoxidase, HppE, is a non-heme-iron peroxidase. Science 342 (2013) 991-995. [PMID: 24114783]
EC 1.11.2.5
Accepted name: 3-methyl-L-tyrosine peroxygenase
Reaction: 3-methyl-L-tyrosine + H2O2 = 3-hydroxy-5-methyl-L-tyrosine + H2O
For diagram of reaction click here.
Other name(s): SfmD; SacD; 3-methyltyrosine peroxidase; 3-methyl-L-tyrosine peroxidase
Systematic name: 3-methyl-L-tyrosine:hydrogen-peroxide oxidoreductase (3-hydroxy-5-methyl-L-tyrosine-forming)
Comments: The heme-containing peroxygenase from the bacterium Streptomyces lavendulae is involved in biosynthesis of saframycin A, a potent antitumor antibiotic that belongs to the tetrahydroisoquinoline family.
References:
1. Tang, M.C., Fu, C.Y. and Tang, G.L. Characterization of SfmD as a heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis. J. Biol. Chem. 287 (2012) 5112-5121. [PMID: 22187429]
EC 1.13.11.78
Accepted name: 2-amino-1-hydroxyethylphosphonate dioxygenase (glycine-forming)
Reaction: (2-amino-1-hydroxyethyl)phosphonate + O2 = glycine + phosphate
Other name(s): phnZ (gene name)
Systematic name: 2-amino-1-hydroxyethylphosphonate:oxygen 1-oxidoreductase (glycine-forming)
Comments: Requires Fe2+. The enzyme, characterized from a marine bacterium, is involved in a 2-aminoethylphosphonate degradation pathway.
References:
1. McSorley, F.R., Wyatt, P.B., Martinez, A., DeLong, E.F., Hove-Jensen, B. and Zechel, D.L. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon-phosphorus bond. J. Am. Chem. Soc. 134 (2012) 8364-8367. [PMID: 22564006]
2. Worsdorfer, B., Lingaraju, M., Yennawar, N.H., Boal, A.K., Krebs, C., Bollinger, J.M., Jr. and Pandelia, M.E. Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Proc. Natl. Acad. Sci. USA 110 (2013) 18874-18879. [PMID: 24198335]
EC 1.13.11.79
Accepted name: 5,6-dimethylbenzimidazole synthase
Reaction: FMNH2 + O2 = 5,6-dimethylbenzimidazole + D-erythrose 4-phosphate + other product(s)
For diagram of reaction click here.
Other name(s): BluB
Systematic name: FMNH2 oxidoreductase (5,6-dimethylbenzimidazole-forming)
Comments: The enzyme catalyses a complex oxygen-dependent conversion of reduced flavin mononucleotide to form 5,6-dimethylbenzimidazole, the lower ligand of vitamin B12. This conversion involves many sequential steps in two distinct stages, and an alloxan intermediate that acts as a proton donor, a proton acceptor, and a hydride acceptor [4]. The C-2 of 5,6-dimethylbenzimidazole is derived from C-1' of the ribityl group of FMNH2 and 2-H from the ribityl 1'-pro-S-hydrogen. While D-erythrose 4-phosphate has been shown to be one of the byproducts, the nature of the other product(s) has not been verified yet.
References:
1. Gray, M.J. and Escalante-Semerena, J.C. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl. Acad. Sci. USA 104 (2007) 2921-2926. [PMID: 17301238]
2. Ealick, S.E. and Begley, T.P. Biochemistry: molecular cannibalism. Nature 446 (2007) 387-388. [PMID: 17377573]
3. Taga, M.E., Larsen, N.A., Howard-Jones, A.R., Walsh, C.T. and Walker, G.C. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446 (2007) 449. [PMID: 17377583]
4. Wang, X.L. and Quan, J.M. Intermediate-assisted multifunctional catalysis in the conversion of flavin to 5,6-dimethylbenzimidazole by BluB: a density functional theory study. J. Am. Chem. Soc. 133 (2011) 4079-4091. [PMID: 21344938]
EC 1.14.11.45
Accepted name: L-isoleucine 4-hydroxylase
Reaction: L-isoleucine + 2-oxoglutarate + O2 = (4S)-4-hydroxy-L-isoleucine + succinate + CO2
Glossary: (4S)-4-hydroxy-L-isoleucine = (2S,3R,4S)-2-amino-4-hydroxy-3-methylpentanoate
Other name(s): ido (gene name)
Systematic name: L-isoleucine,2-oxoglutarate:oxygen oxidoreductase (4-hydroxylating)
Comments: The enzyme, characterized from the bacterium Bacillus thuringiensis, can also catalyse the hydroxylation of L-leucine, L-norvaline, L-norleucine, and L-allo-isoleucine, as well as the sulfoxidation of L-methionine, L-ethionine, S-methyl-L-cysteine, S-ethyl-L-cysteine, and S-allyl-L-cysteine.
References:
1. Kodera, T., Smirnov, S.V., Samsonova, N.N., Kozlov, Y.I., Koyama, R., Hibi, M., Ogawa, J., Yokozeki, K. and Shimizu, S. A novel L-isoleucine hydroxylating enzyme, L-isoleucine dioxygenase from Bacillus thuringiensis, produces (2S,3R,4S)-4-hydroxyisoleucine. Biochem. Biophys. Res. Commun. 390 (2009) 506-510. [PMID: 19850012]
2. Hibi, M., Kawashima, T., Kodera, T., Smirnov, S.V., Sokolov, P.M., Sugiyama, M., Shimizu, S., Yokozeki, K. and Ogawa, J. Characterization of Bacillus thuringiensis-L-isoleucine dioxygenase for production of useful amino acids. Appl. Environ. Microbiol. 77 (2011) 6926-6930. [PMID: 21821743]
3. Hibi, M., Kawashima, T., Yajima, H., Smirnov, S.V., Kodera, T., Sugiyama, M., Shimizu, S., Yokozeki, K., and Ogawa, J. Enzymatic synthesis of chiral amino acid sulfoxides by Fe(II)/α ketoglutarate-dependent dioxygenase. Tetrahedron Asym 24 (2013) 990-994.
EC 1.14.11.46
Accepted name: 2-aminoethylphosphonate dioxygenase
Reaction: (2-aminoethyl)phosphonate + 2-oxoglutarate + O2 = (2-amino-1-hydroxyethyl)phosphonate + succinate + CO2
Other name(s): phnY (gene name)
Systematic name: (2-aminoethyl)phosphonate,2-oxoglutarate:oxygen oxidoreductase (1-hydroxylating)
Comments: Requires Fe2+ and ascorbate. The enzyme, characterized from an uncultured marine bacterium, is involved in a (2-aminoethyl)phosphonate degradation pathway.
References:
1. McSorley, F.R., Wyatt, P.B., Martinez, A., DeLong, E.F., Hove-Jensen, B. and Zechel, D.L. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon-phosphorus bond. J. Am. Chem. Soc. 134 (2012) 8364-8367. [PMID: 22564006]
EC 1.14.13.189
Accepted name: 5-methyl-1-naphthoate 3-hydroxylase
Reaction: 5-methyl-1-naphthoate + NADPH + H+ + O2 = 3-hydroxy-5-methyl-1-naphthoate + NADP+ + H2O
For diagram of reaction click here.
Other name(s): AziB1
Systematic name: 5-methyl-1-naphthoate,NADPH:oxygen oxidoreductase (3-hydroxylating)
Comments: The enzyme from the bacterium Streptomyces sahachiroi is involved in the biosynthesis of 3-methoxy-5-methyl-1-naphthoate, a component of of the the antitumor antibiotic azinomycin B.
References:
1. Ding, W., Deng, W., Tang, M., Zhang, Q., Tang, G., Bi, Y. and Liu, W. Biosynthesis of 3-methoxy-5-methyl naphthoic acid and its incorporation into the antitumor antibiotic azinomycin B. Mol. Biosyst. 6 (2010) 1071-1081. [PMID: 20485749]
EC 1.14.13.190
Accepted name: ferruginol synthase
Reaction: miltiradiene + 2 NADPH + 2 H+ + 2 O2 = ferruginol + 2 NADP+ + 3 H2O
For diagram of reaction click here.
Glossary: miltiradiene = abieta-8,12-diene
Other name(s): miltiradiene oxidase; CYP76AH1
Systematic name: miltiradiene,NADPH:oxygen oxidoreductase (ferruginol forming)
Comments: Isolated from the Chinese medicinal herb Salvia miltiorrhiza (red sage, danshen). It is involved in the biosynthesis of the tanshinones, abietane-type norditerpenoid naphthoquinones that are the main lipophilic bioactive components found in the plant.
References:
1. Guo, J., Zhou, Y.J., Hillwig, M.L., Shen, Y., Yang, L., Wang, Y., Zhang, X., Liu, W., Peters, R.J., Chen, X., Zhao, Z.K. and Huang, L. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 110 (2013) 12108-12113. [PMID: 23812755]
EC 1.14.13.191
Accepted name: ent-sandaracopimaradiene 3-hydroxylase
Reaction: ent-sandaracopimaradiene + NADPH + H+ + O2 = ent-sandaracopimaradien-3β-ol + NADP+ + H2O
For diagram of reaction click here.
Glossary: ent-sandaracopimaradiene = ent-13α-pimara-8(14),15-diene = (4aR,4bR,7S,10aR)-7-ethenyl-1,1,4a,7-tetramethyl-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthrene
Other name(s): CYP701A; OsKOL4
Systematic name: ent-sandaracopimaradiene,NADPH:oxygen oxidoreductase (ent-sandaracopimaradien-3β-ol forming)
Comments: A heme-thiolate protein (cytochrome P-450). Isolated from Oryza sativa (rice). Participates in the pathway for the biosynthesis of oryzalexins, a group of related phytoalexins produced by rice.
References:
1. Wang, Q., Hillwig, M.L., Wu, Y. and Peters, R.J. CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158 (2012) 1418-1425. [PMID: 22247270]
2. Wu, Y., Wang, Q., Hillwig, M.L. and Peters, R.J. Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 454 (2013) 209-216. [PMID: 23795884]
EC 1.14.13.192
Accepted name: oryzalexin E synthase
Reaction: ent-sandaracopimaradien-3β-ol + NADPH + H+ + O2 = oryzalexin E + NADP+ + H2O
For diagram of reaction click here.
Glossary: oryzalexin E = ent-sandaracopimaradiene-3β,9α-diol = (3R,4aR,4bS,7S,10aR)-7-ethenyl-1,1,4a,7-tetramethyl-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthren-2,4b-diol
Other name(s): CYP76M6
Systematic name: ent-sandaracopimaradien-3β-ol,NADPH:oxygen oxidoreductase (oryzalexin E-forming)
Comments: A heme-thiolate protein (cytochrome P-450). Isolated from Oryza sativa (rice). Oryzalexin E is a phytoalexin.
References:
1. Wu, Y., Wang, Q., Hillwig, M.L. and Peters, R.J. Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 454 (2013) 209-216. [PMID: 23795884]
EC 1.14.13.193
Accepted name: oryzalexin D synthase
Reaction: ent-sandaracopimaradien-3β-ol + NADPH + H+ + O2 = oryzalexin D + NADP+ + H2O
For diagram of reaction click here.
Glossary: oryzalexin D = ent-sandaracopimaradiene-3β,7α-diol = (3R,4aR,4bS,7S,9S,10aS)-7-ethenyl-1,1,4a,7-tetramethyl-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthren-2,9-diol
Other name(s): CYP76M8
Systematic name: ent-sandaracopimaradien-3β-ol,NADPH:oxygen oxidoreductase (oryzalexin D forming)
Comments: A heme-thiolate protein (cytochrome P-450). Isolated from Oryza sativa (rice). Oryzalexin D is a phytoalexin.
References:
1. Wu, Y., Wang, Q., Hillwig, M.L. and Peters, R.J. Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 454 (2013) 209-216. [PMID: 23795884]
EC 1.14.13.194
Accepted name: phylloquinone ω-hydroxylase
Reaction: phylloquinone + NADPH + H+ + O2 = ω-hydroxyphylloquinone + NADP+ + H2O
Other name(s): vitamin K1 ω-hydroxylase; CYP4F2; CYP4F11
Systematic name: phylloquinone,NADPH:oxygen oxidoreductase (ω-hydroxyphylloquinone forming)
Comments: A heme-thiolate protein (cytochrome P-450). Isolated from human tissue. The enzyme will also act on menaquinone-4. Prolonged action of CYP4F2, but not CYP4F11, on the ω hydroxyl group oxidizes it to the corresponding carboxylic acid. CYP4F2 also oxidizes leukotriene B4; see EC 1.14.13.30, leukotriene-B4 20-monooxygenase [1].
References:
1. Jin, R., Koop, D.R., Raucy, J.L. and Lasker, J.M. Role of human CYP4F2 in hepatic catabolism of the proinflammatory agent leukotriene B4. Arch. Biochem. Biophys. 359 (1998) 89-98. [PMID: 9799565]
2. Tang, Z., Salamanca-Pinzon, S.G., Wu, Z.L., Xiao, Y. and Guengerich, F.P. Human cytochrome P450 4F11: heterologous expression in bacteria, purification, and characterization of catalytic function. Arch. Biochem. Biophys. 494 (2010) 86-93. [PMID: 19932081]
3. Edson, K.Z., Prasad, B., Unadkat, J.D., Suhara, Y., Okano, T., Guengerich, F.P. and Rettie, A.E. Cytochrome P450-dependent catabolism of vitamin K: ω-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry 52 (2013) 8276-8285. [PMID: 24138531]
EC 1.14.14.15
Accepted name: (3S)-3-amino-3-(3-chloro-4-hydroxyphenyl)propanoyl-[peptidyl-carrier protein SgcC2] monooxygenase
Reaction: (3S)-3-amino-3-(3-chloro-4-hydroxyphenyl)propanoyl-[peptidyl-carrier protein SgcC2] + FADH2 + O2 = (3S)-3-amino-3-(3-chloro-4,5-dihydroxyphenyl)propanoyl-[peptidyl-carrier protein SgcC2] + FAD + H2O
Other name(s): SgcC
Systematic name: (3S)-3-amino-3-(3-chloro-4-hydroxyphenyl)propanoyl-[peptidyl-carrier protein SgcC2],FADH2:oxygen oxidoreductase (5-hydroxylating)
Comments: The enzyme from the actinobacterium Streptomyces globisporus is involved in the biosynthesis of the (S)-3-chloro-5-hydroxy-β-tyrosine moiety prior to incorporation into the chromoprotein antitumor antibiotic C-1027.
References:
1. Lin, S., Van Lanen, S.G. and Shen, B. Characterization of the two-component, FAD-dependent monooxygenase SgcC that requires carrier protein-tethered substrates for the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Am. Chem. Soc. 130 (2008) 6616-6623. [PMID: 18426211]
EC 1.14.16.7
Accepted name: phenylalanine 3-monooxygenase
Reaction: L-phenylalanine + tetrahydrobiopterin + O2 = 3-hydroxy-L-phenylalanine + 4a-hydroxytetrahydrobiopterin
Glossary: 3-hydroxy-L-phenylalanine = meta-L-tyrosine = 3-(3-hydroxyphenyl)-L-alanine
Other name(s): PacX; phenylalanine 3-hydroxylase
Systematic name: L-phenylalanine,tetrahydrobiopterin:oxygen oxidoreductase (3-hydroxylating)
Comments: The enzyme from the bacterium Streptomyces coeruleorubidus forms 3-hydroxy-L-phenylalanine (i.e. m-L-tyrosine), which is one of the building blocks in the biosynthesis of the uridyl peptide antibiotics pacidamycins.
References:
1. Zhang, W., Ames, B.D. and Walsh, C.T. Identification of phenylalanine 3-hydroxylase for meta-tyrosine biosynthesis. Biochemistry 50 (2011) 5401-5403. [PMID: 21615132]
[EC 1.14.19.7 Transferred entry: (S)-2-hydroxypropylphosphonic acid epoxidase. Now EC 1.11.1.23, (S)-2-hydroxypropylphosphonic acid epoxidase. (EC 1.14.19.7 created 2011, deleted 2014)]
[EC 1.14.99.40 Transferred entry: 5,6-dimethylbenzimidazole synthase. Now EC 1.13.11.79, 5,6-dimethylbenzimidazole synthase (EC 1.14.99.40 created 2010, deleted 2014)]
EC 1.16.3.2
Accepted name: bacterial non-heme ferritin
Reaction: 4 Fe(II) + O2 + 6 H2O = 4 [FeO(OH)] + 8 H+ (overall reaction)
Glossary: [FeO(OH)] = iron(III) oxide-hydroxide
Other name(s): FtnA; HuHF
Systematic name: Fe(II):oxygen oxidoreductase ([FeO(OH)]core-producing)
Comments: Ferritins are intracellular iron-storage and detoxification proteins found in all kingdoms of life. They are formed from two subunits that co-assemble in various ratios to form a spherical protein shell. Thousands of mineralized iron atoms are stored within the core of the structure. The product of dioxygen reduction by the bacterial non-heme ferritin is hydrogen peroxide, which is consumed in a subsequent reaction.
References:
1. Hudson, A.J., Andrews, S.C., Hawkins, C., Williams, J.M., Izuhara, M., Meldrum, F.C., Mann, S., Harrison, P.M. and Guest, J.R. Overproduction, purification and characterization of the Escherichia coli ferritin. Eur. J. Biochem. 218 (1993) 985-995. [PMID: 8281950]
2. Stillman, T.J., Hempstead, P.D., Artymiuk, P.J., Andrews, S.C., Hudson, A.J., Treffry, A., Guest, J.R. and Harrison, P.M. The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe3+ and Zn2+ derivatives. J. Mol. Biol. 307 (2001) 587-603. [PMID: 11254384]
3. Bou-Abdallah, F., Yang, H., Awomolo, A., Cooper, B., Woodhall, M.R., Andrews, S.C. and Chasteen, N.D. Functionality of the three-site ferroxidase center of Escherichia coli bacterial ferritin (EcFtnA). Biochemistry 53 (2014) 483-495. [PMID: 24380371]
EC 2.1.1.301
Accepted name: cypemycin N-terminal methyltransferase
Reaction: 2 S-adenosyl-L-methionine + N-terminal L-alanine-[cypemycin] = 2 S-adenosyl-L-homocysteine + N-terminal N,N-dimethyl-L-alanine-[cypemycin]
Other name(s): CypM
Systematic name: S-adenosyl-L-methionine:N-terminal L-alanine-[cypemycin] N-methyltransferase
Comments: The enzyme, isolated from the bacterium Streptomyces sp. OH-4156, can methylate a variety of linear oligopeptides, cyclic peptides such as nisin and haloduracin, and the ε-amino group of lysine [2]. Cypemycin is a peptide antibiotic, a member of the linaridins, a class of posttranslationally modified ribosomally synthesized peptides.
References:
1. Claesen, J. and Bibb, M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc. Natl. Acad. Sci. USA 107 (2010) 16297-16302. [PMID: 20805503]
2. Zhang, Q. and van der Donk, W.A. Catalytic promiscuity of a bacterial α-N-methyltransferase. FEBS Lett 586 (2012) 3391-3397. [PMID: 22841713]
EC 2.1.1.302
Accepted name: 3-hydroxy-5-methyl-1-naphthoate 3-O-methyltransferase
Reaction: S-adenosyl-L-methionine + 3-hydroxy-5-methyl-1-naphthoate = S-adenosyl-L-homocysteine + 3-methoxy-5-methyl-1-naphthoate
For diagram of reaction click here.
Other name(s): AziB2
Systematic name: S-adenosyl-L-methionine:3-hydroxy-5-methyl-1-naphthoate 3-O-methyltransferase
Comments: The enzyme from the bacterium Streptomyces sahachiroi is involved in the biosynthesis of 3-methoxy-5-methyl-1-naphthoate, a component of of the the antitumor antibiotic azinomycin B.
References:
1. Ding, W., Deng, W., Tang, M., Zhang, Q., Tang, G., Bi, Y. and Liu, W. Biosynthesis of 3-methoxy-5-methyl naphthoic acid and its incorporation into the antitumor antibiotic azinomycin B. Mol. Biosyst. 6 (2010) 1071-1081. [PMID: 20485749]
EC 2.1.1.303
Accepted name: 2,7-dihydroxy-5-methyl-1-naphthoate 7-O-methyltransferase
Reaction: S-adenosyl-L-methionine + 2,7-dihydroxy-5-methyl-1-naphthoate = S-adenosyl-L-homocysteine + 2-hydroxy-7-methoxy-5-methyl-1-naphthoate
For diagram of reaction click here.
Other name(s): NcsB1; neocarzinostatin O-methyltransferase
Systematic name: S-adenosyl-L-methionine:2,7-dihydroxy-5-methyl-1-naphthoate 7-O-methyltransferase
Comments: The enzyme from the bacterium Streptomyces carzinostaticus is involved in the biosynthesis of 2-hydroxy-7-methoxy-5-methyl-1-naphthoate. This compound is part of the enediyne chromophore of the antitumor antibiotic neocarzinostatin. In vivo the enzyme catalyses the regiospecific methylation at the 7-hydroxy group of its native substrate 2,7-dihydroxy-5-methyl-1-naphthoate. In vitro it also recognizes other dihydroxynaphthoic acids and catalyses their regiospecific O-methylation.
References:
1. Luo, Y., Lin, S., Zhang, J., Cooke, H.A., Bruner, S.D. and Shen, B. Regiospecific O-methylation of naphthoic acids catalyzed by NcsB1, an O-methyltransferase involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. J. Biol. Chem. 283 (2008) 14694-14702. [PMID: 18387946]
2. Cooke, H.A., Guenther, E.L., Luo, Y., Shen, B. and Bruner, S.D. Molecular basis of substrate promiscuity for the SAM-dependent O-methyltransferase NcsB1, involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. Biochemistry 48 (2009) 9590-9598. [PMID: 19702337]
EC 2.1.1.304
Accepted name: L-tyrosine C3-methyltransferase
Reaction: S-adenosyl-L-methionine + L-tyrosine = S-adenosyl-L-homocysteine + 3-methyl-L-tyrosine
For diagram of reaction click here.
Other name(s): SfmM2; SacF
Systematic name: S-adenosyl-L-methionine:L-tyrosine C3-methyltransferase
Comments: The enzyme from the bacterium Streptomyces lavendulae is involved in biosynthesis of saframycin A, a potent antitumor antibiotic that belongs to the tetrahydroisoquinoline family.
References:
1. Tang, M.C., Fu, C.Y. and Tang, G.L. Characterization of SfmD as a heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis. J. Biol. Chem. 287 (2012) 5112-5121. [PMID: 22187429]
EC 2.1.1.305
Accepted name: 8-demethyl-8-α-L-rhamnosyltetracenomycin-C 2'-O-methyltransferase
Reaction: S-adenosyl-L-methionine + 8-demethyl-8-α-L-rhamnosyltetracenomycin C = S-adenosyl-L-homocysteine + 8-demethyl-8-(2-O-methyl-α-L-rhamnosyl)tetracenomycin C
For diagram of reaction click here.
Glossary: 8-demethyl-8-α-L-rhamnosyltetracenomycin C = methyl (6aR,7S,10aR)-6a,7,10a,12-tetrahydroxy-8-methoxy-1-methyl-6,10,11-trioxo-3-α-L-rhamnosyloxy-6,6a,7,10,10a,11-hexahydrotetracene-2-carboxylate
Other name(s): ElmMI
Systematic name: S-adenosyl-L-methionine:8-demethyl-8-α-L-rhamnosyltetracenomycin-C 2'-O-methyltransferase
Comments: The enzyme from the bacterium Streptomyces olivaceus is involved in the biosynthesis of the polyketide elloramycin.
References:
1. Patallo, E.P., Blanco, G., Fischer, C., Brana, A.F., Rohr, J., Mendez, C. and Salas, J.A. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 276 (2001) 18765-18774. [PMID: 11376004]
EC 2.1.1.306
Accepted name: 8-demethyl-8-(2-methoxy-α-L-rhamnosyl)tetracenomycin-C 3'-O-methyltransferase
Reaction: S-adenosyl-L-methionine + 8-demethyl-8-(2-O-methyl-α-L-rhamnosyl)tetracenomycin C = S-adenosyl-L-homocysteine + 8-demethyl-8-(2,3-di-O-methyl-α-L-rhamnosyl)tetracenomycin C
For diagram of reaction click here.
Glossary: 8-demethyl-8-α-L-rhamnosyltetracenomycin C = methyl (6aR,7S,10aR)-6a,7,10a,12-tetrahydroxy-8-methoxy-1-methyl-6,10,11-trioxo-3-α-L-rhamnosyloxy-6,6a,7,10,10a,11-hexahydrotetracene-2-carboxylate
Other name(s): ElmMII
Systematic name: S-adenosyl-L-methionine:8-demethyl-8-(2-methoxy-α-L-rhamnosyl)tetracenomycin-C 3'-O-methyltransferase
Comments: The enzyme from the bacterium Streptomyces olivaceus is involved in the biosynthesis of the polyketide elloramycin.
References:
1. Patallo, E.P., Blanco, G., Fischer, C., Brana, A.F., Rohr, J., Mendez, C. and Salas, J.A. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 276 (2001) 18765-18774. [PMID: 11376004]
EC 2.1.1.307
Accepted name: 8-demethyl-8-(2,3-dimethoxy-α-L-rhamnosyl)tetracenomycin-C 4'-O-methyltransferase
Reaction: S-adenosyl-L-methionine + 8-demethyl-8-(2,3-di-O-methyl-α-L-rhamnosyl)tetracenomycin C = S-adenosyl-L-homocysteine + 8-demethyl-8-(2,3,4-tri-O-methyl-α-L-rhamnosyl)tetracenomycin C
For diagram of reaction click here.
Glossary: 8-demethyl-8-α-L-rhamnosyltetracenomycin C = methyl (6aR,7S,10aR)-6a,7,10a,12-tetrahydroxy-8-methoxy-1-methyl-6,10,11-trioxo-3-α-L-rhamnosyloxy-6,6a,7,10,10a,11-hexahydrotetracene-2-carboxylate
Other name(s): ElmMIII
Systematic name: S-adenosyl-L-methionine:8-demethyl-8-(2,3-di-O-methoxy-α-L-rhamnosyl)tetracenomycin-C 4'-O-methyltransferase
Comments: The enzyme from the bacterium Streptomyces olivaceus is involved in the biosynthesis of the polyketide elloramycin.
References:
1. Patallo, E.P., Blanco, G., Fischer, C., Brana, A.F., Rohr, J., Mendez, C. and Salas, J.A. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 276 (2001) 18765-18774. [PMID: 11376004]
[EC 2.1.3.13 Deleted entry: ATP carbamoyltransferase. The enzyme has been replaced by EC 6.1.2.2, nebramycin 5' synthase. (EC 2.1.3.13 created 2013, deleted 2014)]
[EC 2.1.3.14 Deleted entry: tobramycin carbamoyltransferase. The enzyme has been replaced by EC 6.1.2.2, nebramycin 5' synthase (EC 2.1.3.14 created 2013, deleted 2014)]
EC 2.3.1.233
Accepted name: 1,3,6,8-tetrahydroxynaphthalene synthase
Reaction: 5 malonyl-CoA = 1,3,6,8-tetrahydroxynaphthalene + 5 CoA + 5 CO2 + H2O
For diagram of reaction click here.
Other name(s): PKS1; THNS; SCO1206; RppA
Systematic name: malonyl-CoA C-acyl transferase (1,3,6,8-tetrahydroxynaphthalene forming)
Comments: Isolated from the fungus Colletotrichum lagenarium [1], and the bacteria Streptomyces coelicolor [2,3] and Streptomyces peucetius [4]. It only uses malonyl-CoA, without invovement of acetyl-CoA.
References:
1. Fujii, I., Mori, Y., Watanabe, A., Kubo, Y., Tsuji, G. and Ebizuka, Y. Enzymatic synthesis of 1,3,6,8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase PKS1. Biochemistry 39 (2000) 8853-8858. [PMID: 10913297]
2. Izumikawa, M., Shipley, P.R., Hopke, J.N., O'Hare, T., Xiang, L., Noel, J.P. and Moore, B.S. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 30 (2003) 510-515. [PMID: 12905073]
3. Austin, M.B., Izumikawa, M., Bowman, M.E., Udwary, D.W., Ferrer, J.L., Moore, B.S. and Noel, J.P. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J. Biol. Chem. 279 (2004) 45162-45174. [PMID: 15265863]
4. Ghimire, G.P., Oh, T.J., Liou, K. and Sohng, J.K. Identification of a cryptic type III polyketide synthase (1,3,6,8-tetrahydroxynaphthalene synthase) from Streptomyces peucetius ATCC 27952. Mol. Cells 26 (2008) 362-367. [PMID: 18612244]
*EC 2.4.1.208
Accepted name: diglucosyl diacylglycerol synthase (1,2-linking)
Reaction: UDP-α-D-glucose + 1,2-diacyl-3-O-(α-D-glucopyranosyl)-sn-glycerol = 1,2-diacyl-3-O-[α-D-glucopyranosyl-(1→2)-O-α-D-glucopyranosyl]-sn-glycerol + UDP
Other name(s): monoglucosyl diacylglycerol (1→2) glucosyltransferase; MGlcDAG (1→2) glucosyltransferase; DGlcDAG synthase (ambiguous); UDP-glucose:1,2-diacyl-3-O-(α-D-glucopyranosyl)-sn-glycerol (1→2) glucosyltransferase; diglucosyl diacylglycerol synthase
Systematic name: UDP-α-D-glucose:1,2-diacyl-3-O-(α-D-glucopyranosyl)-sn-glycerol 2-glucosyltransferase
Comments: The enzyme from Acholeplasma laidlawii requires Mg2+.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number: 168680-19-1
References:
1. Karlsson, O.P., Rytomaa, M., Dahlqvist, A., Kinnunen, P.K., Wieslander, A. Correlation between bilayer lipid dynamics and activity of the diglucosyldiacylglycerol synthase from Acholeplasma laidlawii membranes. Biochemistry 35 (1996) 10094-10102. [PMID: 8756472]
EC 2.4.1.325
Accepted name: TDP-N-acetylfucosamine:lipid II N-acetylfucosaminyltransferase
Reaction: dTDP-4-acetamido-4,6-dideoxy-α-D-galactose + N-acetyl-β-D-mannosaminouronyl-(1→4)-N-acetyl-α-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol = dTDP + 4-acetamido-4,6-dideoxy-α-D-galactosyl-(1→4)-N-acetyl-β-D-mannosaminouronyl-(1→4)-N-acetyl-α-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
Glossary: dTDP-4-acetamido-4,6-dideoxy-α-D-galactose = dTDP-N-acetyl-α-D-fucosamine
Other name(s): TDP-Fuc4NAc:lipid II Fuc4NAc-transferase; TDP-Fuc4NAc:lipid II Fuc4NAc transferase; wecF (gene name)
Systematic name: dTDP-N-acetyl-α-D-fucose:N-acetyl-β-D-mannosaminouronyl-(1→4)-N-acetyl-α-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol N-acetylfucosaminyltransferase
Comments: Involved in the enterobacterial common antigen (ECA) biosynthesis in the bacterium Escherichia coli. The trisaccharide of the product (lipid III) is the repeat unit of ECA.
References:
1. Rahman, A., Barr, K. and Rick, P.D. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 183 (2001) 6509-6516. [PMID: 11673418]
EC 2.4.1.326
Accepted name: aklavinone 7-β-L-rhodosaminyltransferase
Reaction: dTDP-β-L-rhodosamine + aklavinone = dTDP + aclacinomycin T
For diagram of reaction click here.
Glossary: dTDP-β-L-rhodosamine = dTDP-2,3,6-trideoxy-3-dimethylamino-β-L-lyxo-hexose
Other name(s): AknS/AknT
Systematic name: dTDP-β-L-rhodosamine:aklavinone 7-α-L-rhodosaminyltransferase
Comments: Isolated from the bacterium Streptomyces galilaeus. Forms a complex with its accessory protein AknT, and has very low activity in its absence. The enzyme can also use dTDP-2-deoxy-β-L-fucose. Involved in the biosynthesis of other aclacinomycins.
References:
1. Lu, W., Leimkuhler, C., Gatto, G.J., Jr., Kruger, R.G., Oberthur, M., Kahne, D. and Walsh, C.T. AknT is an activating protein for the glycosyltransferase AknS in L-aminodeoxysugar transfer to the aglycone of aclacinomycin A. Chem. Biol. 12 (2005) 527-534. [PMID: 15911373]
2. Leimkuhler, C., Fridman, M., Lupoli, T., Walker, S., Walsh, C.T. and Kahne, D. Characterization of rhodosaminyl transfer by the AknS/AknT glycosylation complex and its use in reconstituting the biosynthetic pathway of aclacinomycin A. J. Am. Chem. Soc. 129 (2007) 10546-10550. [PMID: 17685523]
EC 2.4.1.327
Accepted name: aclacinomycin-T 2-deoxy-L-fucose transferase
Reaction: dTDP-2-deoxy-β-L-fucose + aclacinomycin T = dTDP + aclacinomycin S
For diagram of reaction click here.
Glossary: idarubicin = (7S,9S)-9-acetyl-7-(3-amino-2,3,6-trideoxy-β-L-lyxo-hexosyloxy)-6,9,11-trihydroxy-7,8,9,10-tetrahydrotetracene-5,12-dione
Other name(s): AknK
Systematic name: dTDP-2-deoxy-β-L-fucose:7-(α-L-rhodosaminyl)aklavinone 2-deoxy-α-L-fucosyltransferase
Comments: The enzyme, isolated from the bacterium Streptomyces galilaeus, is involved in the biosynthesis of other aclacinomycins. Also acts on idarubicin. It will slowly add a second 2-deoxy-L-fucose unit to aclacinomycin S in vitro.
References:
1. Lu, W., Leimkuhler, C., Oberthur, M., Kahne, D. and Walsh, C.T. AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A. Biochemistry 43 (2004) 4548-4558. [PMID: 15078101]
EC 2.4.1.328
Accepted name: erythronolide mycarosyltransferase
Reaction: dTDP-β-L-mycarose + erythronolide B = dTDP + 3-α-L-mycarosylerythronolide B
For diagram of reaction click here.
Glossary: dTDP-β-L-mycarose = dTDP-2,6-dideoxy-3-C-methyl-β-L-ribo-hexose
Other name(s): EryBV
Systematic name: dTDP-β-L-mycarose:erythronolide B L-mycarosyltransferase
Comments: Isolated from the bacterium Saccharopolyspora erythraea. The enzyme is involved in the biosynthesis of the antibiotic erythromycin.
References:
1. Zhang, C., Fu, Q., Albermann, C., Li, L. and Thorson, J.S. The in vitro characterization of the erythronolide mycarosyltransferase EryBV and its utility in macrolide diversification. Chembiochem 8 (2007) 385-390. [PMID: 17262863]
EC 2.5.1.121
Accepted name: 5,10-dihydrophenazine-1-carboxylate 9-dimethylallyltransferase
Reaction: dimethylallyl diphosphate + 5,10-dihydrophenazine-1-carboxylate = diphosphate + 9-(dimethylallyl)-5,10-dihydrophenazine-1-carboxylate
For diagram of reaction click here.
Glossary: 9-(dimethylallyl)-5,10-dihydrophenazine-1-carboxylate = 9-(3-methylbut-2-en-1-yl)-5,10-dihydrophenazine-1-carboxylate
Other name(s): PpzP; dihydrophenazine-1-carboxylate dimethylallyltransferase; 5,10-dihydrophenazine 1-carboxylate dimethylallyltransferase
Systematic name: dimethylallyl diphosphate:5,10-dihydrophenazine-1-carboxylate 9-dimethylallyltransferase
Comments: The enzyme is involved in the biosynthesis of prenylated phenazines by the bacterium Streptomyces anulatus. It is specific for both dimethylallyl diphosphate and 5,10-dihydrophenazine-1-carboxylate.
References:
1. Saleh, O., Gust, B., Boll, B., Fiedler, H.P. and Heide, L. Aromatic prenylation in phenazine biosynthesis: dihydrophenazine-1-carboxylate dimethylallyltransferase from Streptomyces anulatus. J. Biol. Chem. 284 (2009) 14439-14447. [PMID: 19339241]
EC 2.5.1.122
Accepted name: 4-O-dimethylallyl-L-tyrosine synthase
Reaction: dimethylallyl diphosphate + L-tyrosine = diphosphate + 4-O-dimethylallyl-L-tyrosine
Other name(s): SirD
Systematic name: dimethylallyl diphosphate:L-tyrosine 4-O-dimethylallyltransferase
Comments: The enzyme is involved in biosynthesis of the phytotoxin sirodesmin PL by the phytopathogenic ascomycete Leptosphaeria maculans.
References:
1. Kremer, A. and Li, S.M. A tyrosine O-prenyltransferase catalyses the first pathway-specific step in the biosynthesis of sirodesmin PL. Microbiology 156 (2010) 278-286. [PMID: 19762440]
2. Zou, H.X., Xie, X., Zheng, X.D. and Li, S.M. The tyrosine O-prenyltransferase SirD catalyzes O-, N-, and C-prenylations. Appl. Microbiol. Biotechnol. 89 (2011) 1443-1451. [PMID: 21038099]
EC 2.5.1.123
Accepted name: flaviolin linalyltransferase
Reaction: geranyl diphosphate + flaviolin = 3-linalylflaviolin + diphosphate
For diagram of reaction click here.
Glossary: flaviolin = 2,5,7-trihydroxynaphthalene-1,4-dione
Other name(s): Fnq26
Systematic name: geranyl-diphosphate:flaviolin 3-linalyltransferase
Comments: Does not require Mg2+ or any other metal ions. Isolated from the bacterium Streptomyces cinnamonensis. In vitro the enzyme also forms traces of 3-geranylflaviolin.
References:
1. Haagen, Y., Unsold, I., Westrich, L., Gust, B., Richard, S.B., Noel, J.P. and Heide, L. A soluble, magnesium-independent prenyltransferase catalyzes reverse and regular C-prenylations and O-prenylations of aromatic substrates. FEBS Lett 581 (2007) 2889-2893. [PMID: 17543953]
EC 2.7.1.183
Accepted name: glycoprotein-mannosyl O6-kinase
Reaction: ATP + 3-O-[N-acetyl-β-D-galactosaminyl-(1→3)-N-acetyl-β-D-glucosaminyl-(1→4)-α-D-mannosyl]-L-threonyl/L-seryl-[protein] = ADP + 3-O-[N-acetyl-β-D-galactosaminyl-(1→3)-N-acetyl-β-D-glucosaminyl-(1→4)-6-O-phosphono-α-D-mannosyl]-L-threonyl/L-seryl-[protein]
For diagram of reaction click here.
Other name(s): SGK196; protein O-mannose kinase
Systematic name: ATP:3-O-[N-acetyl-β-D-galactosaminyl-(1→3)-N-acetyl-β-D-glucosaminyl-(1→4)-α-D-mannosyl]-L-threonyl/L-seryl-[protein] 6-phosphotransferase
Comments: In humans this phosphorylated trisaccharide is attached to an L-threonine residue of α-dystroglycan, an extracellular peripheral glycoprotein that acts as a receptor for extracellular matrix proteins containing laminin-G domains, and is important for its activity.
References:
1. Yoshida-Moriguchi, T., Willer, T., Anderson, M.E., Venzke, D., Whyte, T., Muntoni, F., Lee, H., Nelson, S.F., Yu, L. and Campbell, K.P. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341 (2013) 896-899. [PMID: 23929950]
EC 2.7.1.184
Accepted name: sulfofructose kinase
Reaction: ATP + 6-deoxy-6-sulfo-D-fructose = ADP + 6-deoxy-6-sulfo-D-fructose 1-phosphate
For diagram of reaction click here.
Other name(s): yihV (gene name)
Systematic name: ATP:6-deoxy-6-sulfo-D-fructose 1-phosphotransferase
Comments: The enzyme, characterized from the bacterium Escherichia coli, is involved in the degradation pathway of sulfoquinovose, the polar headgroup of sulfolipids found in the photosynthetic membranes of all higher plants, mosses, ferns, algae, and most photosynthetic bacteria, as well as the surface layer of some archaea.
References:
1. Denger, K., Weiss, M., Felux, A.K., Schneider, A., Mayer, C., Spiteller, D., Huhn, T., Cook, A.M. and Schleheck, D. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507 (2014) 114-117. [PMID: 24463506]
*EC 2.7.7.86
Accepted name: cyclic GMP-AMP synthase
Reaction: ATP + GTP = 2 diphosphate + cyclic Gp(2'-5')Ap(3'-5') (overall reaction)
Glossary: cyclic Gp(2'-5')Ap(3'-5') = cyclo[(3'→5')-guanylyl-(2'→5')-adenylyl]
Other name(s): cGAMP synthase; cGAS
Systematic name: ATP:GTP adenylyltransferase (cyclizing)
Comments: Cyclic Gp(2'-5')Ap(3'-5') is a signalling molecule in mammalian cells that triggers the production of type I interferons and other cytokines.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number:
References:
1. Sun, L., Wu, J., Du, F., Chen, X. and Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339 (2013) 786-791. [PMID: 23258413]
2. Ablasser, A., Goldeck, M., Cavlar, T., Deimling, T., Witte, G., Rohl, I., Hopfner, K.P., Ludwig, J. and Hornung, V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature 498 (2013) 380-384. [PMID: 23722158]
EC 2.8.2.37
Accepted name: trehalose 2-sulfotransferase
Reaction: 3'-phosphoadenylyl sulfate + α,α-trehalose = adenosine 3',5'-bisphosphate + 2-O-sulfo-α,α-trehalose
Glossary: 2-O-sulfo-α,α-trehalose = trehalose 2-sulfate = α-D-glucopyranosyl 2-O-sulfo-α-D-glucopyranoside
Other name(s): Stf0 sulfotransferase
Systematic name: 3'-phosphoadenylyl-sulfate:α,α-trehalose 2-sulfotransferase
Comments: The sulfation of trehalose in the bacterium Mycobacterium tuberculosis is required for the biosynthesis of sulfolipid-1.
References:
1. Mougous, J.D., Petzold, C.J., Senaratne, R.H., Lee, D.H., Akey, D.L., Lin, F.L., Munchel, S.E., Pratt, M.R., Riley, L.W., Leary, J.A., Berger, J.M. and Bertozzi, C.R. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat. Struct. Mol. Biol. 11 (2004) 721-729. [PMID: 15258569]
2. Pi, N., Hoang, M.B., Gao, H., Mougous, J.D., Bertozzi, C.R. and Leary, J.A. Kinetic measurements and mechanism determination of Stf0 sulfotransferase using mass spectrometry. Anal. Biochem. 341 (2005) 94-104. [PMID: 15866533]
EC 3.1.1.96
Accepted name: D-aminoacyl-tRNA deacylase
Reaction: a D-aminoacyl-tRNA + H2O = a D-amino acid + tRNA
Other name(s): Dtd2; D-Tyr-tRNA(Tyr) deacylase; D-Tyr-tRNATyr deacylase; D-tyrosyl-tRNATyr aminoacylhydrolase; dtdA (gene name)
Systematic name: D-aminoacyl-tRNA aminoacylhydrolase
Comments: The enzyme from Escherichia coli can cleave D-tyrosyl-tRNATyr, D-aspartyl-tRNAAsp and D-tryptophanyl-tRNATrp [1]. Whereas the enzyme from the archaeon Pyrococcus abyssi is a zinc protein, the enzyme from Escherichia coli does not carry any zinc [2].
References:
1. Soutourina, J., Plateau, P. and Blanquet, S. Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem. 275 (2000) 32535-32542. [PMID: 10918062]
2. Ferri-Fioni, M.L., Schmitt, E., Soutourina, J., Plateau, P., Mechulam, Y. and Blanquet, S. Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases. J. Biol. Chem. 276 (2001) 47285-47290. [PMID: 11568181]
3. Ferri-Fioni, M.L., Fromant, M., Bouin, A.P., Aubard, C., Lazennec, C., Plateau, P. and Blanquet, S. Identification in archaea of a novel D-Tyr-tRNATyr deacylase. J. Biol. Chem. 281 (2006) 27575-27585. [PMID: 16844682]
4. Wydau, S., Ferri-Fioni, M.L., Blanquet, S. and Plateau, P. GEK1, a gene product of Arabidopsis thaliana involved in ethanol tolerance, is a D-aminoacyl-tRNA deacylase. Nucleic Acids Res. 35 (2007) 930-938. [PMID: 17251192]
EC 3.1.4.57
Accepted name: phosphoribosyl 1,2-cyclic phosphate 1,2-diphosphodiesterase
Reaction: (1) 5-phospho-α-D-ribose 1,2-cyclic phosphate + H2O = D-ribofuranose 2,5-bisphosphate
Other name(s): cyclic phosphate dihydrolase; phnPP (gene name)
Systematic name: 5-phospho-α-D-ribose 1,2-cyclic phosphate 1,2-diphosphophosphohydrolase
Comments: The enzyme, characterized from the bacterium Eggerthella lenta, is involed in degradation of methylphosphonate.
References:
1. Ghodge, S.V., Cummings, J.A., Williams, H.J. and Raushel, F.M. Discovery of a cyclic phosphodiesterase that catalyzes the sequential hydrolysis of both ester bonds to phosphorus. J. Am. Chem. Soc. 135 (2013) 16360-16363. [PMID: 24147537]
EC 3.1.12 Exodeoxyribonucleases producing 3'-phosphomonoesters
EC 3.1.12.1
Accepted name: 5' to 3' exodeoxyribonuclease (nucleoside 3'-phosphate-forming)
Reaction: exonucleolytic cleavage in the 5'- to 3'-direction to yield nucleoside 3'-phosphates
Other name(s): Cas4; 5' to 3' single stranded DNA exonuclease
Comments: Preference for single-stranded DNA. The enzyme from the archaeon Sulfolobus solfataricus contains a [4Fe-4S] cluster and requires a divalent metal cation, such as Mg2+ or Mn2+, for activity.
References:
1. Zhang, J., Kasciukovic, T. and White, M.F. The CRISPR associated protein Cas4 Is a 5' to 3' DNA exonuclease with an iron-sulfur cluster. PLoS One 7 (2012) e47232. [PMID: 23056615]
2. Lemak, S., Beloglazova, N., Nocek, B., Skarina, T., Flick, R., Brown, G., Popovic, A., Joachimiak, A., Savchenko, A. and Yakunin, A.F. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S]-cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 135 (2013) 17476-17487. [PMID: 24171432]
*EC 3.5.1.46
Accepted name: 6-aminohexanoate-oligomer exohydrolase
Reaction: (1) [N-(6-aminohexanoyl)]n + H2O = [N-(6-aminohexanoyl)]n-1 + 6-aminohexanoate
Other name(s): 6-aminohexanoate-dimer hydrolase; nylB (gene name); 6-aminohexanoic acid oligomer hydrolase (ambiguous); N-(6-aminohexanoyl)-6-aminohexanoate amidohydrolase; nylon-6 hydrolase (ambiguous)
Systematic name: N-(6-aminohexanoyl)-6-aminohexanoate exoamidohydrolase
Comments: The enzyme is involved in degradation of nylon-6 oligomers. It degrades linear oligomers of 6-aminohexanoate with a degree of polymerization of 2-20 by exo-type cleavage, removing residues sequentially from the N-terminus. Activity decreases with the increase of the polymerization number of the oligomer. cf. EC 3.5.1.117, 6-aminohexanoate-oligomer endohydrolase and EC 3.5.2.12, 6-aminohexanoate-cyclic-dimer hydrolase.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
PDB,
UM-BBD,
CAS registry number: 75216-15-8
References:
1. Kinoshita, S., Terada, T., Taniguchi, T., Takeney, Y., Masuda, S., Matsunaga, N. and Okada, H. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. KI72. Eur. J. Biochem. 116 (1981) 547-551. [PMID: 7262074]
EC 3.5.1.117
Accepted name: 6-aminohexanoate-oligomer endohydrolase
Reaction: [N-(6-aminohexanoyl)]n + H2O = [N-(6-aminohexanoyl)]n-x + [N-(6-aminohexanoyl)]x
Other name(s): endo-type 6-aminohexanoate oligomer hydrolase; Ahx endo-type-oligomer hydrolase; 6-aminohexanoate oligomer hydrolase; Ahx-oligomer hydrolase; nylon hydrolase; nylon-oligomer hydrolase; NylC; nylon-6 hydrolase (ambiguous)
Systematic name: 6-aminohexanoate oligomer endoamidohydrolase
Comments: The enzyme is involved in degradation of nylon-6 oligomers. It degrades linear or cyclic oligomers of poly(6-aminohexanoate) with a degree of polymerization greater than three (n-> 3) by endo-type cleavage, to oligomers of a length of two or more (2 ≤ x-< n). It shows negligible activity with N-(6-aminohexanoyl)-6-aminohexanoate (cf. EC 3.5.1.46, 6-aminohexanoate-oligomer exo hydrolase) or with 1,8-diazacyclotetradecane-2,9-dione (cf. EC 3.5.2.12, 6-aminohexanoate-cyclic-dimer hydrolase).
References:
1. Kakudo, S., Negoro, S., Urabe, I. and Okada, H. Nylon oligomer degradation gene, nylC, on plasmid pOAD2 from a Flavobacterium strain encodes endo-type 6-aminohexanoate oligomer hydrolase: purification and characterization of the nylC gene product. Appl. Environ. Microbiol. 59 (1993) 3978-3980. [PMID: 8285701]
2. Yasuhira, K., Tanaka, Y., Shibata, H., Kawashima, Y., Ohara, A., Kato, D., Takeo, M. and Negoro, S. 6-Aminohexanoate oligomer hydrolases from the alkalophilic bacteria Agromyces sp. strain KY5R and Kocuria sp. strain KY2. Appl. Environ. Microbiol. 73 (2007) 7099-7102. [PMID: 17827307]
3. Negoro, S., Shibata, N., Tanaka, Y., Yasuhira, K., Shibata, H., Hashimoto, H., Lee, Y.H., Oshima, S., Santa, R., Oshima, S., Mochiji, K., Goto, Y., Ikegami, T., Nagai, K., Kato, D., Takeo, M. and Higuchi, Y. Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis. J. Biol. Chem. 287 (2012) 5079-5090. [PMID: 22187439]
*EC 3.5.4.17
Accepted name: adenosine-phosphate deaminase
Reaction: (1) AMP + H2O = IMP + NH3
Glossary: IMP = inosine 5'-phosphate
Other name(s): adenylate deaminase; adenine nucleotide deaminase; adenosine (phosphate) deaminase
Systematic name: adenosine-phosphate aminohydrolase
Comments: Acts on AMP, ADP, ATP, NAD+ and adenosine, in decreasing order of activity. The bacterial enzyme can also accept the deoxy derivatives. cf. EC 3.5.4.6, AMP deaminase.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number: 37289-20-6
References:
1. Su, J.-C., Li, C.-C. and Ting, C.C. A new adenylate deaminase from red marine alga Porphyra crispata. Biochemistry 5 (1966) 536-543. [PMID: 5940938]
2. Yates, M.G. A non-specific adenine nucleotide deaminase from Desulfovibrio desulfuricans. Biochim. Biophys. Acta 171 (1969) 299-310. [PMID: 5773435]
*EC 3.5.99.7
Accepted name: 1-aminocyclopropane-1-carboxylate deaminase
Reaction: 1-aminocyclopropane-1-carboxylate + H2O = 2-oxobutanoate + NH3 (overall reaction)
Other name(s): 1-aminocyclopropane-1-carboxylate endolyase (deaminating); ACC deaminase; 1-aminocyclopropane carboxylic acid deaminase
Systematic name: 1-aminocyclopropane-1-carboxylate aminohydrolase (isomerizing)
Comments: A pyridoxal 5'-phosphate enzyme. The enzyme, found in certain soil bacteria and fungi, catalyses the ring opening of 1-aminocyclopropane-1-carboxylate, the immediate precursor to ethylene, an important plant hormone that regulates fruit ripening and other processes. The enzyme releases an unstable enamine product that tautomerizes to an imine form, which undergoes a hydrolytic deamination. The latter reaction, which can occur spontaneously, can also be catalysed by EC 3.5.99.10, 2-iminobutanoate/2-iminopropanoate deaminase. The enzyme has been used to make fruit ripening dependent on externally added ethylene, as it removes the substrate for endogenous ethylene formation.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
PDB,
UM-BBD,
CAS registry number: 69553-48-6
References:
1. Honma, M. and Shimomura, T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42 (1978) 1825-1831.
2. Yao, M., Ose, T., Sugimoto, H., Horiuchi, A., Nakagawa, A., Wakatsuki, S., Yokoi, D., Murakami, T., Honma, M. and Tanaka, I. Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus. J. Biol. Chem. 275 (2000) 34557-34565. [PMID: 10938279]
3. Thibodeaux, C.J. and Liu, H.W. Mechanistic studies of 1-aminocyclopropane-1-carboxylate deaminase: characterization of an unusual pyridoxal 5'-phosphate-dependent reaction. Biochemistry 50 (2011) 1950-1962. [PMID: 21244019]
EC 3.13.1.4
Accepted name: 3-sulfinopropanoyl-CoA desulfinase
Reaction: 3-sulfinopropanoyl-CoA + H2O = propanoyl-CoA + sulfite
Other name(s): 3SP-CoA desulfinase; AcdDPN7; 3-sulfinopropionyl-CoA desulfinase
Systematic name: 3-sulfinopropanoyl-CoA sulfinohydrolase
Comments: The enzyme from the β-proteobacterium Advenella mimigardefordensis contains one non-covalently bound FAD per subunit.
References:
1. Schurmann, M., Deters, A., Wubbeler, J.H. and Steinbuchel, A. A novel 3-sulfinopropionyl coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis strain DPN7T acting as a key enzyme during catabolism of 3,3'-dithiodipropionic acid is a member of the acyl-CoA dehydrogenase superfamily. J. Bacteriol. 195 (2013) 1538-1551. [PMID: 23354747]
2. Schurmann, M., Demming, R.M., Krewing, M., Rose, J., Wubbeler, J.H. and Steinbuchel, A. Identification of 3-sulfinopropionyl coenzyme A (CoA) desulfinases within the Acyl-CoA dehydrogenase superfamily. J. Bacteriol. 196 (2014) 882-893. [PMID: 24317404]
EC 4.1.1.98
Accepted name: 4-hydroxy-3-polyprenylbenzoate decarboxylase
Reaction: a 4-hydroxy-3-polyprenylbenzoate = a 2-polyprenylphenol + CO2
Other name(s): ubiX (gene name); ubiD (gene name); slr1099 (gene name); PAD1 (gene name); 4-hydroxy-3-solanesylbenzoate decarboxylase; 3-octaprenyl-4-hydroxybenzoate decarboxylase
Systematic name: 4-hydroxy-3-polyprenylbenzoate carboxy-lyase
Comments: The enzyme catalyses a step in prokaryotic ubiquinone biosynthesis, as well as in plastoquinone biosynthesis in cyanobacteria. The enzyme can accept substrates with different polyprenyl tail lengths in vitro, but uses a specific length in vivo, which is determined by the polyprenyl diphosphate synthase that exists in the specific organism.
References:
1. Leppik, R.A., Young, I.G. and Gibson, F. Membrane-associated reactions in ubiquinone biosynthesis in Escherichia coli. 3-Octaprenyl-4-hydroxybenzoate carboxy-lyase. Biochim. Biophys. Acta 436 (1976) 800-810. [PMID: 782527]
2. Gulmezian, M., Hyman, K.R., Marbois, B.N., Clarke, C.F. and Javor, G.T. The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467 (2007) 144-153. [PMID: 17889824]
3. Pfaff, C., Glindemann, N., Gruber, J., Frentzen, M. and Sadre, R. Chorismate pyruvate-lyase and 4-hydroxy-3-solanesylbenzoate decarboxylase are required for plastoquinone biosynthesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. (2013) . [PMID: 24337576]
EC 4.1.2.57
Accepted name: sulfofructosephosphate aldolase
Reaction: 6-deoxy-6-sulfo-D-fructose 1-phosphate = glycerone phosphate + 2-hydroxy-3-oxopropane-1-sulfonate
For diagram of reaction click here.
Glossary: glycerone phosphate = dihydroxyacetone phosphate = 3-hydroxy-2-oxopropyl phosphate
Other name(s): yihT (gene name)
Systematic name: 6-deoxy-6-sulfofructose-1-phosphate 2-hydroxy-3-oxopropane-1-sulfonate-lyase (glycerone-phosphate-forming)
Comments: The enzyme, characterized from the bacterium Escherichia coli, is involved in the degradation pathway of sulfoquinovose, the polar headgroup of sulfolipids found in the photosynthetic membranes of all higher plants, mosses, ferns, algae, and most photosynthetic bacteria, as well as the surface layer of some archaea.
References:
1. Denger, K., Weiss, M., Felux, A.K., Schneider, A., Mayer, C., Spiteller, D., Huhn, T., Cook, A.M. and Schleheck, D. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507 (2014) 114-117. [PMID: 24463506]
*EC 4.2.1.134
Accepted name: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase
Reaction: a very-long-chain (3R)-3-hydroxyacyl-CoA = a very-long-chain trans-2,3-dehydroacyl-CoA + H2O
Glossary: a very-long-chain acyl-CoA = an acyl-CoA thioester where the acyl chain contains 23 or more carbon atoms.
Other name(s): PHS1 (gene name); PAS2 (gene name)
Systematic name: very-long-chain (3R)-3-hydroxyacyl-CoA hydro-lyase
Comments: This is the third component of the elongase, a microsomal protein complex responsible for extending palmitoyl-CoA and stearoyl-CoA (and modified forms thereof) to very-long chain acyl CoAs. cf. EC 2.3.1.199, very-long-chain 3-oxoacyl-CoA synthase, EC 1.1.1.330, very-long-chain 3-oxoacyl-CoA reductase, and EC 1.3.1.93, very-long-chain enoyl-CoA reductase.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number:
References:
1. Bach, L., Michaelson, L.V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., Da Costa, M., Boutin, J.P., Miquel, M., Tellier, F., Domergue, F., Markham, J.E., Beaudoin, F., Napier, J.A. and Faure, J.D. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 105 (2008) 14727-14731. [PMID: 18799749]
2. Kihara, A., Sakuraba, H., Ikeda, M., Denpoh, A. and Igarashi, Y. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J. Biol. Chem. 283 (2008) 11199-11209. [PMID: 18272525]
EC 4.2.1.152
Accepted name: hydroperoxy icosatetraenoate dehydratase
Reaction: a hydroperoxyicosatetraenoate = an oxoicosatetraenoate + H2O
Glossary: (12R)-HPETE = (5Z,8Z,10E,12R,14Z)-12-hydroperoxyicosa-5,8,10,14-tetraenoate
Other name(s): epidermal lipoxygenase-3 (ambiguous); eLOX3 (ambiguous)
Systematic name: hydroperoxyicosatetraenoate hydro-lyase (oxoicosatetraenoate-forming)
Comments: Binds Fe2+. The mammalian enzymes accept a range of hydroperoxyicosatetraenoates (HPETE). The human enzyme has highest activity with (12R)-HPETE, followed by (12S)-HPETE and (15R)-HPETE with much lower efficiency. The murine enzyme has highest activity with (8R)-HPETE followed by (8S)-HPETE. All HPETE isoforms are converted to the corresponding oxoicosatetraenoate forms (KETE) [2]. The enzymes also catalyse the reaction of EC 5.4.4.7, hydroperoxy icosatetraenoate isomerase.
References:
1. Yu, Z., Schneider, C., Boeglin, W.E., Marnett, L.J. and Brash, A.R. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc. Natl. Acad. Sci. USA 100 (2003) 9162-9167. [PMID: 12881489]
2. Yu, Z., Schneider, C., Boeglin, W.E. and Brash, A.R. Human and mouse eLOX3 have distinct substrate specificities: implications for their linkage with lipoxygenases in skin. Arch. Biochem. Biophys. 455 (2006) 188-196. [PMID: 17045234]
3. Zheng, Y. and Brash, A.R. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J. Biol. Chem. 285 (2010) 39866-39875. [PMID: 20921226]
EC 4.2.1.153
Accepted name: 3-methylfumaryl-CoA hydratase
Reaction: (S)-citramalyl-CoA = 3-methylfumaryl-CoA + H2O
For diagram of reaction click here.
Glossary: (S)-citramalyl-CoA = (3S)-3-carboxy-3-hydroxybutanoyl-CoA
Other name(s): Meh; mesaconyl-C4-CoA hydratase; mesaconyl-coenzyme A hydratase (ambiguous)
Systematic name: (S)-citramalyl-CoA hydro-lyase (3-methylfumaryl-CoA-forming)
Comments: The enzyme from the bacterium Chloroflexus aurantiacus is part of the 3-hydroxypropanoate cycle for carbon assimilation.
References:
1. Zarzycki, J., Brecht, V., Muller, M. and Fuchs, G. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc. Natl. Acad. Sci. USA 106 (2009) 21317-21322. [PMID: 19955419]
EC 4.2.1.154
Accepted name: tetracenomycin F2 cyclase
Reaction: tetracenomycin F2 = tetracenomycin F1 + H2O
For diagram of reaction click here.
Glossary: tetracenomycin F1 = 3,8,10,12-tetrahydroxy-1-methyl-11-oxo-6,11-dihydro-2-tetracenecarboxylate = 6,11-dihydro-3,8,10,12-tetrahydroxy-1-methyl-11-oxonaphthacene-2-carboxylate
Other name(s): tcmI (gene name)
Systematic name: tetracenomycin F2 hydro-lyase (tetracenomycin-F1-forming)
Comments: The enzyme is involved in biosynthesis of the anthracycline antibiotic tetracenomycin C by the bacterium Streptomyces glaucescens.
References:
1. Shen, B. and Hutchinson, C.R. Tetracenomycin F2 cyclase: intramolecular aldol condensation in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32 (1993) 11149-11154. [PMID: 8218177]
2. Thompson, T.B., Katayama, K., Watanabe, K., Hutchinson, C.R. and Rayment, I. Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase. J. Biol. Chem. 279 (2004) 37956-37963. [PMID: 15231835]
EC 4.2.3.145
Accepted name: ophiobolin F synthase
Reaction: (2E,6E,10E,14E)-geranylfarnesyl diphosphate + H2O = ophiobolin F + diphosphate
For diagram of reaction click here.
Systematic name: (2E,6E,10E,14E)-geranylfarnesyl-diphosphate diphosphate-lyase (cyclizing, ophiobolin-F-forming)
Comments: Isolated from the fungus Aspergillus clavatus. The product is a sesterterpenoid (C25 terpenoid).
References:
1. Chiba, R., Minami, A., Gomi, K. and Oikawa, H. Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus. Org. Lett. 15 (2013) 594-597. [PMID: 23324037]
EC 4.2.3.146
Accepted name: cyclooctat-9-en-7-ol synthase
Reaction: geranylgeranyl diphosphate + H2O = cyclooctat-9-en-7-ol + diphosphate
For diagram of reaction click here and mechanism click here.
Glossary: cyclooctat-9-en-7-ol = (1S,3aS,4R,7S,9aS,10aS)-1,4,9a-trimethyl-7-(propan-2-yl)-1,2,3,3a,4,5,7,8,9,9a,10,10a-dodecahydrodicyclopenta[a,d][8]annulen-4-ol
Other name(s): cetB2
Systematic name: geranylgeranyl-diphosphate diphosphate-lyase (cyclooctat-9-en-7-ol-forming)
Comments: Requires Mg2+. Isolated from the bacterium Streptomyces melanosporofaciens, where it is part of the biosynthesis of cyclooctatin, a potent inhibitor of lysophospholipase.
References:
1. Kim, S.Y., Zhao, P., Igarashi, M., Sawa, R., Tomita, T., Nishiyama, M. and Kuzuyama, T. Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene cyclase and two P450 hydroxylases. Chem. Biol. 16 (2009) 736-743. [PMID: 19635410]
2. Zhang, X., Shang, G., Gu, L. and Shen, Y. Crystallization and preliminary X-ray diffraction analysis of the diterpene cyclooctatin synthase (CYC) from Streptomyces sp. LZ35. Acta Crystallogr. F Struct. Biol. Commun. 70 (2014) 366-369. [PMID: 24598929]
EC 5.1.3.29
Accepted name: L-fucose mutarotase
Reaction: α-L-fucopyranose = β-L-fucopyranose
Other name(s): FucU; fucose mutarotase; FucM
Systematic name: L-fucose 1-epimerase
Comments: This enzyme shows no 1-epimerase activity with D-glucose, L-rhamnose and D-fucose (cf. EC 5.1.3.3, aldose 1-epimerase) [1].
References:
1. Ryu, K.S., Kim, C., Kim, I., Yoo, S., Choi, B.S. and Park, C. NMR application probes a novel and ubiquitous family of enzymes that alter monosaccharide configuration. J. Biol. Chem. 279 (2004) 25544-25548. [PMID: 15060078]
2. Park, D., Ryu, K.S., Choi, D., Kwak, J. and Park, C. Characterization and role of fucose mutarotase in mammalian cells. Glycobiology 17 (2007) 955-962. [PMID: 17602138]
EC 5.3.1.31
Accepted name: sulfoquinovose isomerase
Reaction: sulfoquinovose = 6-deoxy-6-sulfo-D-fructose
For diagram of reaction click here.
Glossary: sulfoquinovose = 6-deoxy-6-sulfo-D-glucopyranose
Other name(s): yihS (gene name)
Systematic name: 6-deoxy-6-sulfo-D-glucopyranose aldose-ketose-isomerase
Comments: The enzyme, characterized from the bacterium Escherichia coli, is involved in the degradation pathway of sulfoquinovose, the polar headgroup of sulfolipids found in the photosynthetic membranes of all higher plants, mosses, ferns, algae, and most photosynthetic bacteria, as well as the surface layer of some archaea.
References:
1. Denger, K., Weiss, M., Felux, A.K., Schneider, A., Mayer, C., Spiteller, D., Huhn, T., Cook, A.M. and Schleheck, D. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507 (2014) 114-117. [PMID: 24463506]
*EC 5.4.2.11
Accepted name: phosphoglycerate mutase (2,3-diphosphoglycerate-dependent)
Reaction: 2-phospho-D-glycerate = 3-phospho-D-glycerate (overall reaction)
For diagram of reaction click here.
Glossary: 2/3-phospho-D-glycerate = 2-phospho-D-glycerate or 3-phospho-D-glycerate
Other name(s): glycerate phosphomutase (diphosphoglycerate cofactor); 2,3-diphosphoglycerate dependent phosphoglycerate mutase; cofactor dependent phosphoglycerate mutase; phosphoglycerate phosphomutase (ambiguous); phosphoglyceromutase (ambiguous); monophosphoglycerate mutase (ambiguous); monophosphoglyceromutase (ambiguous); GriP mutase (ambiguous); PGA mutase (ambiguous); MPGM; PGAM; PGAM-d; PGM; dPGM
Systematic name: D-phosphoglycerate 2,3-phosphomutase (2,3-diphosphoglycerate-dependent)
Comments: The enzymes from vertebrates, platyhelminths, mollusks, annelids, crustaceans, insects, algae, fungi, yeast and some bacteria (particularly Gram-negative) require 2,3-bisphospho-D-glycerate as a cofactor. The enzyme is activated by 2,3-bisphospho-D-glycerate by transferring a phosphate to histidine (His10 in man and Escherichia coli, His8 in Saccharomyces cerevisiae). This phosphate can be transferred to the free OH of 2-phospho-D-glycerate, followed by transfer of the phosphate already on the phosphoglycerate back to the histidine. cf. EC 5.4.2.12 phosphoglycerate mutase. The enzyme has no requirement for metal ions. This enzyme also catalyse, slowly, the reactions of EC 5.4.2.4 bisphosphoglycerate mutase.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc
References:
1. Grisolia, S. Phosphoglyceric acid mutase. Methods Enzymol. 5 (1962) 236-242.
2. Ray, W.J., Jr. and Peck, E.J., Jr. Phosphomutases. In: Boyer, P.D. (Ed.), The Enzymes, 3rd edn, vol. 6, 1972, pp. 407-477.
3. Rose, Z.B. The enzymology of 2,3-bisphosphoglycerate. Adv. Enzymol. Relat. Areas Mol. Biol. 51 (1980) 211-253. [PMID: 6255773]
4. Rigden, D.J., Walter, R.A., Phillips, S.E. and Fothergill-Gilmore, L.A. Sulphate ions observed in the 2.12 Å structure of a new crystal form of S. cerevisiae phosphoglycerate mutase provide insights into understanding the catalytic mechanism. J. Mol. Biol. 286 (1999) 1507-1517. [PMID: 10064712]
5. Bond, C.S., White, M.F. and Hunter, W.N. High resolution structure of the phosphohistidine-activated form of Escherichia coli cofactor-dependent phosphoglycerate mutase. J. Biol. Chem. 276 (2001) 3247-3253. [PMID: 11038361]
6. Rigden, D.J., Mello, L.V., Setlow, P. and Jedrzejas, M.J. Structure and mechanism of action of a cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus with broad specificity phosphatase activity. J. Mol. Biol. 315 (2002) 1129-1143. [PMID: 11827481]
7. Rigden, D.J., Littlejohn, J.E., Henderson, K. and Jedrzejas, M.J. Structures of phosphate and trivanadate complexes of Bacillus stearothermophilus phosphatase PhoE: structural and functional analysis in the cofactor-dependent phosphoglycerate mutase superfamily. J. Mol. Biol. 325 (2003) 411-420. [PMID: 12498792]
EC 5.4.4.7
Accepted name: hydroperoxy icosatetraenoate isomerase
Reaction: a hydroperoxyicosatetraenoate = a hydroxyepoxyicosatrienoate
Glossary: (12R)-HPETE = (5Z,8Z,10E,12R,14Z)-12-hydroperoxyicosa-5,8,10,14-tetraenoate
Other name(s): epidermal lipoxygenase-3 (ambiguous); eLOX3 (ambiguous)
Systematic name: hydroperoxyicosatetraenoate hydroxymutase
Comments: Binds Fe2+. The enzyme from mammals accepts a range of hydroperoxyicosatetraenoates producing one or several different hydroxyepoxyicosatrienoates. The human enzyme has highest activity with (12R)-HPETE producing (5Z,8R,9E,11R,12R,14Z)-8-hydroxy-11,12-epoxyicosa-5,9,14-trienoate, followed by (12S)-HPETE producing (5Z,8Z,10R,11S,12S,14Z)-10-hydroxy-11,12-epoxyicosa-5,8,14-trienoate and (5Z,8R,9E,11S,12S,14Z)-8-hydroxy-11,12-epoxyicosa-5,9,14-trienoate [1]. The mouse enzyme has highest activity with (8S)-HPETE, producing (5Z,8S,9S,10R,11Z,14Z)-10-hydroxy-8,9-epoxyicosa-5,11,14-trienoate [2]. The enzymes also have the activity of EC 4.2.1.152, hydroperoxy icosatetraenoate dehydratase.
References:
1. Yu, Z., Schneider, C., Boeglin, W.E., Marnett, L.J. and Brash, A.R. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc. Natl. Acad. Sci. USA 100 (2003) 9162-9167. [PMID: 12881489]
2. Yu, Z., Schneider, C., Boeglin, W.E. and Brash, A.R. Human and mouse eLOX3 have distinct substrate specificities: implications for their linkage with lipoxygenases in skin. Arch. Biochem. Biophys. 455 (2006) 188-196. [PMID: 17045234]
3. Zheng, Y. and Brash, A.R. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J. Biol. Chem. 285 (2010) 39866-39875. [PMID: 20921226]
*EC 5.5.1.23
Accepted name: aklanonic acid methyl ester cyclase
Reaction: aklaviketone = methyl aklanonate
For diagram of reaction click here.
Glossary: aklaviketone = methyl (1R,2R)-2-ethyl-2,5,7-trihydroxy-4,6,11-trioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate
Other name(s): dauD (gene name); aknH (gene name); dnrD (gene name); methyl aklanonate cyclase; methyl aklanonate-aklaviketone isomerase (cyclizing)
Systematic name: aklaviketone lyase (decyclizing)
Comments: The enzyme is involved in the biosynthesis of aklaviketone, an intermediate in the biosynthetic pathways leading to formation of several anthracycline antibiotics, including aclacinomycin, daunorubicin and doxorubicin.
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number:
References:
1. Dickens, M.L., Ye, J. and Strohl, W.R. Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomyces sp. strain C5. J. Bacteriol. 177 (1995) 536-543. [PMID: 7836284]
2. Kendrew, S.G., Katayama, K., Deutsch, E., Madduri, K. and Hutchinson, C.R. DnrD cyclase involved in the biosynthesis of doxorubicin: purification and characterization of the recombinant enzyme. Biochemistry 38 (1999) 4794-4799. [PMID: 10200167]
3. Kallio, P., Sultana, A., Niemi, J., Mantsala, P. and Schneider, G. Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. J. Mol. Biol. 357 (2006) 210-220. [PMID: 16414075]
EC 6.1.2.2
Accepted name: nebramycin 5' synthase
Reaction: (1) tobramycin + carbamoyl phosphate + ATP + H2O = nebramycin 5' + AMP + diphosphate + phosphate (overall reaction)
For diagram of reaction click here.
Glossary: tobramycin = (1S,2S,3R,4S,6R)-4,6-diamino-3-(2,6-diamino-2,3,6-trideoxy-α-D-ribo-hexopyranosyloxy)-2-hydroxycyclohexyl 3-amino-3-deoxy-α-D-glucopyranoside
Other name(s): tobramycin carbamoyltransferase; TobZ
Systematic name: tobramycin:carbamoyl phosphate ligase (AMP,phosphate-forming)
Comments: Requires Fe(III). The enzyme from the bacterium Streptoalloteichus tenebrarius catalyses the activation of carbamoyl phosphate to O-carbamoyladenylate and the subsequent carbamoylation of kanamycin and tobramycin.
References:
1. Parthier, C., Gorlich, S., Jaenecke, F., Breithaupt, C., Brauer, U., Fandrich, U., Clausnitzer, D., Wehmeier, U.F., Bottcher, C., Scheel, D. and Stubbs, M.T. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew. Chem. Int. Ed. Engl. 51 (2012) 4046-4052. [PMID: 22383337]
EC 6.2.1.41
Accepted name: 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoate—CoA ligase
Reaction: ATP + 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoate + CoA = AMP + diphosphate + 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoyl-CoA
For diagram of reaction click here.
Glossary: 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoate = HIP
Other name(s): fadD3 (gene name); HIP—CoA ligase
Systematic name: 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoate:CoA ligase (AMP-forming)
Comments: The enzyme, characterized from actinobacterium Mycobacterium tuberculosis, catalyses a step in the degradation of cholesterol and cholate. The enzyme is very specific for its substrate, and requires that the side chain at C17 is completely removed.
References:
1. Horinouchi, M., Hayashi, T., Koshino, H. and Kudo, T. ORF18-disrupted mutant of Comamonas testosteroni TA441 accumulates significant amounts of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and its derivatives after incubation with steroids. J. Steroid Biochem. Mol. Biol. 101 (2006) 78-84. [PMID: 16891113]
2. Casabon, I., Crowe, A.M., Liu, J. and Eltis, L.D. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol. Microbiol. 87 (2013) 269-283. [PMID: 23146019]
EC 6.2.1.42
Accepted name: 3-oxocholest-4-en-26-oate—CoA ligase
Reaction: ATP + 3-oxocholest-4-en-26-oate + CoA = AMP + diphosphate + 3-oxocholest-4-en-26-oyl-CoA
For diagram of reaction click here.
Other name(s): fadD19 (gene name)
Systematic name: 3-oxocholest-4-en-26-oate:CoA ligase (AMP-forming)
Comments: The enzyme, characterized from actinobacterium Mycobacterium tuberculosis, catalyses a step in the degradation of cholesterol. It is responsible for the activation of the C8 side chain. 3β-hydroxycholest-5-en-26-oate can also be used as substrate.
References:
1. Wilbrink, M.H., Petrusma, M., Dijkhuizen, L. and van der Geize, R. FadD19 of Rhodococcus rhodochrous-DSM43269, a steroid-coenzyme A ligase essential for degradation of C-24 branched sterol side chains. Appl. Environ. Microbiol. 77 (2011) 4455-4464. [PMID: 21602385]
2. Casabon, I., Swain, K., Crowe, A.M., Eltis, L.D. and Mohn, W.W. Actinobacterial acyl coenzyme a synthetases involved in steroid side-chain catabolism. J. Bacteriol. 196 (2014) 579-587. [PMID: 24244004]
EC 6.2.1.43
Accepted name: 2-hydroxy-7-methoxy-5-methyl-1-naphthoate—CoA ligase
Reaction: ATP + 2-hydroxy-7-methoxy-5-methyl-1-naphthoate + CoA = AMP + diphosphate + 2-hydroxy-7-methoxy-5-methyl-1-naphthoyl-CoA
For diagram of reaction click here.
Other name(s): NcsB2
Systematic name: 2-hydroxy-7-methoxy-5-methyl-1-naphthoate:CoA ligase
Comments: The enzyme from the bacterium Streptomyces carzinostaticus is involved in the attachment of the 2-hydroxy-7-methoxy-5-methyl-1-naphthoate moiety of the antibiotic neocarzinostatin. In vitro the enzyme also catalyses the activation of other 1-naphthoic acid analogues, e.g. 2-hydroxy-5-methyl-1-naphthoate or 2,7-dihydroxy-5-methyl-1-naphthoate.
References:
1. Cooke, H.A., Zhang, J., Griffin, M.A., Nonaka, K., Van Lanen, S.G., Shen, B. and Bruner, S.D. Characterization of NcsB2 as a promiscuous naphthoic acid/coenzyme A ligase integral to the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. J. Am. Chem. Soc. 129 (2007) 7728-7729. [PMID: 17539640]
*EC 6.3.1.9
Accepted name: trypanothione synthase
Reaction: (1) glutathione + spermidine + ATP = glutathionylspermidine + ADP + phosphate
For diagram of reaction click here.
Glossary: N1,N8-bis(glutathionyl)spermidine = trypanothione
Other name(s): glutathionylspermidine:glutathione ligase (ADP-forming)
Systematic name: spermidine/glutathionylspermidine:glutathione ligase (ADP-forming)
Comments: The enzyme, characterized from several trypanosomatids (e.g. Trypanosoma cruzi) catalyses two subsequent reactions, leading to production of trypanothione from glutathione and spermidine. Some trypanosomatids (e.g. Crithidia species and some Leishmania species) also contain an enzyme that only carries out the first reaction (cf. EC 6.3.1.8, glutathionylspermidine synthase). The enzyme is bifunctional, and also catalyses the hydrolysis of glutathionylspermidine and trypanothione (cf. EC 3.5.1.78, glutathionylspermidine amidase).
Links to other databases:
BRENDA,
EXPASY,
KEGG,
MetaCyc,
CAS registry number: 130246-69-4
References:
1. Smith, K., Nadeau, K., Bradley, M., Walsh, C.T., Fairlamb, A.H. Purification of glutathionylspermidine and trypanothione synthase from Crithidia fasciculata. Protein Sci. 1 (1992) 874-883. [PMID: 1304372]
2. Oza, S.L., Tetaud, E., Ariyanayagam, M.R., Warnon, S.S. and Fairlamb, A.H. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in Trypanosoma cruzi. J. Biol. Chem. 277 (2002) 35853-35861. [PMID: 12121990]
3. Comini, M., Menge, U., Wissing, J. and Flohe, L. Trypanothione synthesis in crithidia revisited. J. Biol. Chem. 280 (2005) 6850-6860. [PMID: 15537651]
4. Oza, S.L., Shaw, M.P., Wyllie, S. and Fairlamb, A.H. Trypanothione biosynthesis in Leishmania major. Mol. Biochem. Parasitol. 139 (2005) 107-116. [PMID: 15610825]
5. Fyfe, P.K., Oza, S.L., Fairlamb, A.H. and Hunter, W.N. Leishmania trypanothione synthetase-amidase structure reveals a basis for regulation of conflicting synthetic and hydrolytic activities. J. Biol. Chem. 283 (2008) 17672-17680. [PMID: 18420578]
EC 6.3.1.18
Accepted name: γ-glutamylanilide synthase
Reaction: ATP + L-glutamate + aniline = ADP + phosphate + N5-phenyl-L-glutamine
Glossary: γ-glutamylanilide = N5-phenyl-L-glutamine
Other name(s): atdA1 (gene name); tdnQ (gene name); dcaQ (gene name)
Systematic name: L-glutamate:aniline ligase (ADP-forming)
Comments: Requires Mg2+. The enzyme, characterized from the bacterium Acinetobacter sp. YAA, catalyses the first step in the degradation of aniline. It can also accept chlorinated and methylated forms of aniline, preferrably in the o- and p-positions.
References:
1. Takeo, M., Ohara, A., Sakae, S., Okamoto, Y., Kitamura, C., Kato, D. and Negoro, S. Function of a glutamine synthetase-like protein in bacterial aniline oxidation via γ-glutamylanilide. J. Bacteriol. 195 (2013) 4406-4414. [PMID: 23893114]
2-(4-hydroxyphenyl)-2-oxoacetate = 4-hydroxyphenylglyoxylate = (4-hydroxyphenyl)(oxo)acetate
L-(4-hydroxyphenyl)glycine = (S)-4-hydroxyphenylglycine
L-(3,5-dihydroxyphenyl)glycine = (S)-3,5-dihydroxyphenylglycine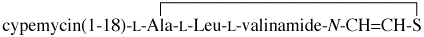
ferruginol = abieta-8,11,13-trien-12-ol
ent-sandaracopimaradien-3β-ol = (3R,4aR,4bR,7S,10aS)-7-ethenyl-1,1,4a,7-tetramethyl-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthren-2-ol
ent-sandaracopimaradien-3β-ol = (3R,4aR,4bR,7S,10aS)-7-ethenyl-1,1,4a,7-tetramethyl-1,2,3,4,4a,4b,5,6,7,9,10,10a-dodecahydrophenanthren-2-ol
(1a) 2 Fe(II) + O2 + 4 H2O = 2 [FeO(OH)] + 4 H+ + H2O2
(1b) 2 Fe(II) + H2O2 + 2 H2O = 2 [FeO(OH)] + 4 H+
lipid II = N-acetyl-β-D-mannosaminouronyl-(1→4)-N-acetyl-α-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
lipid III = N-acetyl-β-D-fucosyl-(1→4)-N-acetyl-β-D-mannosaminouronyl-(1→4)-N-acetyl-α-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
aklavinone = methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate
aclacinomycin T = 7-O-(α-L-rhodosaminyl)aklavinone
aclacinomycin S = 7-O-(2-deoxy-α-L-fucosyl-(1→4)-rhodosaminyl)aklavinone
aclacinomycin T = 7-O-(α-L-rhodosaminyl)aklavinone
L-mycarose = 2,6-dideoxy-3-C-methyl-L-ribo-hexose
3-linalylflaviolin = 2,5,7-trihydroxy-3-(3,7-dimethylocta-1,6-dien-3-yl)naphthalene-1,4-dione
(1a) ATP + GTP = pppGp(2'-5')A + diphosphate
(1b) pppGp(2'-5')A = cyclic Gp(2'-5')Ap(3'-5') + diphosphate
(2) D-ribofuranose 2,5-bisphosphate + H2O = D-ribofuranose 5-phosphate + phosphate
(2) N-(6-aminohexanoyl)-6-aminohexanoate + H2O = 2 6-aminohexanoate
(2) ADP + H2O = IDP + NH3
(3) ATP + H2O = ITP + NH3
AMP = adenosine 5'-phosphate
(1a) 1-aminocyclopropane-1-carboxylate = 2-aminobut-2-enoate
(1b) 2-aminobut-2-enoate = 2-iminobutanoate (spontaneous)
(1c) 2-iminobutanoate + H2O = 2-oxobutanoate + NH3 (spontaneous)
2-hydroxy-3-oxopropane-1-sulfonate = 3-sulfolactaldehyde
(12S)-HPETE = (5Z,8Z,10E,12S,14Z)-12-hydroperoxyicosa-5,8,10,14-tetraenoate
12-KETE = 12-oxo-ETE = (5Z,8Z,10E,14Z)-12-oxoicosa-5,8,10,14-tetraenoate
(8R)-HPETE = (5Z,8R,9E,11Z,14Z)-8-hydroperoxyicosa-5,9,11,14-tetraenoate
(15R)-HPETE = (5Z,8Z,11Z,13E,15R)-15-hydroperoxyicosa-5,8,11,13-tetraenoate
3-methylfumaryl-CoA = (E)-3-carboxybut-2-enoyl-CoA
tetracenomycin F2 = (3E)-4-(3-acetyl-4,5,7-trihydroxy-10-oxo-9,10-dihydroanthracen-2-yl)-3-hydroxybut-3-enoate
(1a) [enzyme]-L-histidine + 2,3-bisphospho-D-glycerate = [enzyme]-Nτ-phospho-L-histidine + 2/3-phospho-D-glycerate
(1b) [enzyme]-Nτ-phospho-L-histidine + 2-phospho-D-glycerate = [enzyme]-L-histidine + 2,3-bisphospho-D-glycerate
(1c) [enzyme]-L-histidine + 2,3-bisphospho-D-glycerate = [enzyme]-Nτ-phospho-L-histidine + 3-phospho-D-glycerate
(1d) [enzyme]-Nτ-phospho-L-histidine + 2/3-bisphospho-D-glycerate = [enzyme]-L-histidine + 2,3-bisphospho-D-glycerate
(8S)-HPETE = (5Z,8S,9E,11Z,14Z)-8-hydroperoxyicosa-5,9,11,14-tetraenoate
methyl aklanonate = methyl [4,5-dihydroxy-9,10-dioxo-3-(3-oxopentanoyl)-9,10-dihydroanthracen-2-yl]acetate
(1a) carbamoyl phosphate + ATP + H2O = diphosphate + O-carbamoyladenylate + phosphate
(1b) O-carbamoyladenylate + tobramycin = AMP + nebramycin 5'
(2) kanamycin A + carbamoyl phosphate + ATP + H2O = 6''-O-carbamoylkanamycin A + AMP + diphosphate + phosphate (overall reaction)
(2a) carbamoyl phosphate + ATP + H2O = diphosphate + O-carbamoyladenylate + phosphate
(2b) O-carbamoyladenylate + kanamycin A = AMP + 6''-O-carbamoylkanamycin A
nebramycin 5' = (1S,2S,3R,4S,6R)-4,6-diamino-3-[(2,6-diamino-2,3,6-trideoxy-α-D-ribo-hexopyranosyl)oxy]-2-hydroxycyclohexyl 3-amino-6-O-carbamoyl-3-deoxy-α-D-glucopyranoside
kanamycin A = (1S,2R,3R,4S,6R)-4,6-diamino-3-(6-amino-6-deoxy--D-glucopyranosyloxy)-2-hydroxycyclohexyl 3-amino-3-deoxy--D-glucopyranoside
6''-O-carbamoylkanamycin A = (1S,2R,3R,4S,6R)-4,6-diamino-3-[(6-amino-6-deoxy-α-D-glucopyranosyl)oxy]-2-hydroxycyclohexyl 3-amino-6-O-carbamoyl-3-deoxy-α-D-glucopyranoside
(2) glutathione + glutathionylspermidine + ATP = N1,N8-bis(glutathionyl)spermidine + ADP + phosphate
Return to enzymes home page.